- RNA-II Lab, Department of Biochemical Engineering and Biotechnology, Indian Institute of Technology Delhi, New Delhi, India
Specific microRNAs have emerged as key players in disease biology by playing crucial role in disease development and progression. This review draws attention to one such microRNA, miR-191 that has been recently reported to be abnormally expressed in several cancers (>20) and various other diseases like diabetes-type 2, Crohn’s, pulmonary hypertension, and Alzheimer’s. It regulates important cellular processes such as cell proliferation, differentiation, apoptosis, and migration by targeting important transcription factors, chromatin remodelers, and cell cycle associated genes. Several studies have demonstrated it to be an excellent biomarker for cancer diagnosis and prognosis leading to two patents already in its kitty. In this first review we summarize the current knowledge of the regulation, functions and targets of miR-191 and discuss its potential as a promising disease biomarker and therapeutic target.
Introduction
MicroRNAs are a class of small non-coding RNAs that function as post-transcriptional gene regulators (Bartel and Chen, 2004). Volinia et al. (2006) for the first time suggested the association of solid tumors with dysregulation of specific microRNAs. Since then miRNA-cancer link has extended beyond reports of abnormal miRNA levels in various cancers to miRNAs acting themselves as oncomiRs or tumor-suppressors. However, functional significance of majority of dysregulated miRNAs has not been elucidated. We have focused here on one such highly conserved miRNA, miR-191 that was found to be abnormally expressed in more than twenty different cancers and shown to be a major player in some of these. Further, altered expression of miR-191 has also been associated with various other diseases such as type 2 diabetes, neurodegenerative diseases, and more recently with innate immunity (Saba et al., 2008; Xu et al., 2009; Zampetaki et al., 2010). This review is timely to highlight the crucial role miR-191 plays in a multitude of diseases including cancer and also in normal development and differentiation processes. We start off with a small introduction to miR-191/425 cluster and its regulation followed by summarizing studies on functional dissection of miR-191 in various cancers. Overall, current understanding of miR-191 biology suggests it to be a promising novel target for cancer prognosis and therapy.
miR-191/425 Cluster
miR-191 is expressed as part of the miR-191/425 cluster which is highly conserved in several metazoan species (miR-425 in 26 species, miR-191 in 30 species) suggesting it to be an important player in higher eukaryotes (Kiezun et al., 2012). First discovered in mouse, its expression was later confirmed in humans in a leukemia cell line (HL-60) and subsequently in 20 different human tissues (Lagos-Quintana et al., 2003; Kasashima et al., 2004). This cluster is located in the first intron of DALRD3 gene on human chromosome 3 (3p21.31) and codes for four mature miRNAs – miR-191-5p, miR-191-3p, miR-425-5p, and miR-425-3p. It forms a sense antisense transcript pair with DALRD3, NDUFA3, and WDR6 mRNAs; however, the functional impact of this association has not been experimentally evaluated (Griffiths-Jones et al., 2006). miR-191 tends to be coexpressed with the DALRD3 and NDUFAF3 transcripts. miR-191 resides 381 nucleotides upstream of miR-425 and notably shows much higher expression or cloning frequency as compared to miR-425 in all the tissues studied so far (Altuvia et al., 2005; Di Leva et al., 2013). This is in compliance with recent reports showing that huge variations in expression levels of miRNA members may exist within the same cluster (Vaz et al., 2010). Indeed, Croce and Kulshreshthas group have shown that both miR-191 and miR-425 promote cell proliferation in breast cancer exhibiting overlapping functions, but a previous study shows that miR-191 and not miR-425 promotes erythroid enucleation (Zhang et al., 2011; Di Leva et al., 2013; Nagpal et al., 2013). Therefore, with very limited data, we assume that miR-191 and miR-425 may exhibit specific or overlapping functions depending on the tissue and conditions.
miR-191 in Development and Differentiation
The first report that miR-191 is developmentally regulated came from the observation that significantly high levels of miR-191 were present in (postmortem) human prefrontal cortex specimens of individuals older than 41 years than those under 15 years. It was suggested that miR-191 mediated downregulation of brain-derived neurotrophic factor (BDNF; predicted miR-191 target) may contribute to the cortical development (Mellios et al., 2008). miR-191 is also one of the major miRNA expressed in rat neurons and its expression increases ~2–5 fold with time in rat cortical cultures suggestive of its role in neuronal development (Kim et al., 2004).
Developmental association of miR-191 was further extended to senescence since its overexpression was shown to promote replicative senescence and inhibit proliferation in primary human keratinocytes (Lena et al., 2012). Role of miR-191 in regulation of circadian rhythm was also suggested as it was shown to exhibit a circadian pattern of expression in mouse liver and showed inverse correlation to Bma-1 (circadian transcription factor) levels (Na et al., 2009).
The connection of miR-191 and differentiation was also explored recently. miR-191 was found to be significantly downregulated during terminal erythroid differentiation (CFU-E to the Ter119+ stage) while its overexpression inhibited erythroid enucleation and chromatin condensation suggesting that miR-191 plays a crucial role in erythropoiesis (Zhang et al., 2011). Levels of miR-191 were also found to be differentially expressed in monocytes vs. monocytes derived dendritic cells (Cekaite et al., 2010). Induced miR-191 levels were observed in osteoblast like cells following treatment with Bio-Oss (an organic bovine bone). As Bio-Oss plays an important role in bone regeneration thus miR-191 may be an important mediator for the same (Palmieri et al., 2010). Therefore, all these findings suggest that miR-191 plays an important role in cellular differentiation and development.
miR-191 and Cancer Connection
miR-191 and cancer abnormalities have been reported in more than twenty different malignancies making the miR-191 a ubiquitously notorious miRNA like miR-21 or miR-155. Overexpression of miR-191 in 16 different cancer types [breast (female), colon, lung, liver, prostate, pancreas, stomach, ovarian cancer, pituitary adenoma, esophageal squamous carcinoma, oral squamous carcinoma, osteosarcoma, B-ALL, bladder, anaplastic large cell lymphoma, and acute myeloid leukemia (AML)] classifies it largely as an oncogenic miRNA. However, in six other cancer types (severe medulloblastomas, retinoblastoma, thyroid follicular tumor, male breast cancer, CALL, and melanoma) its levels are known to be downregulated (Volinia et al., 2006; Xi et al., 2006; Garzon et al., 2008; Fassan et al., 2009; Ferretti et al., 2009; Fulci et al., 2009; Hui et al., 2009; Kent et al., 2009; Caramuta et al., 2010; Elyakim et al., 2010; Patnaik et al., 2010; Shen et al., 2010; Colamaio et al., 2011; Duan et al., 2011; He et al., 2011; Leite et al., 2011, 2013; Li et al., 2011; Shi et al., 2011; Xu et al., 2011; Inoue et al., 2012; McEvoy et al., 2012; Poliseno et al., 2012; Di Leva et al., 2013; Gombos et al., 2013; Liu et al., 2013a; Nagpal et al., 2013; Zhou et al., 2013; Scheffer et al., 2014). The nature of miR-191 deregulation, its functions and targets reported so far in various cancers are shown in Table 1.
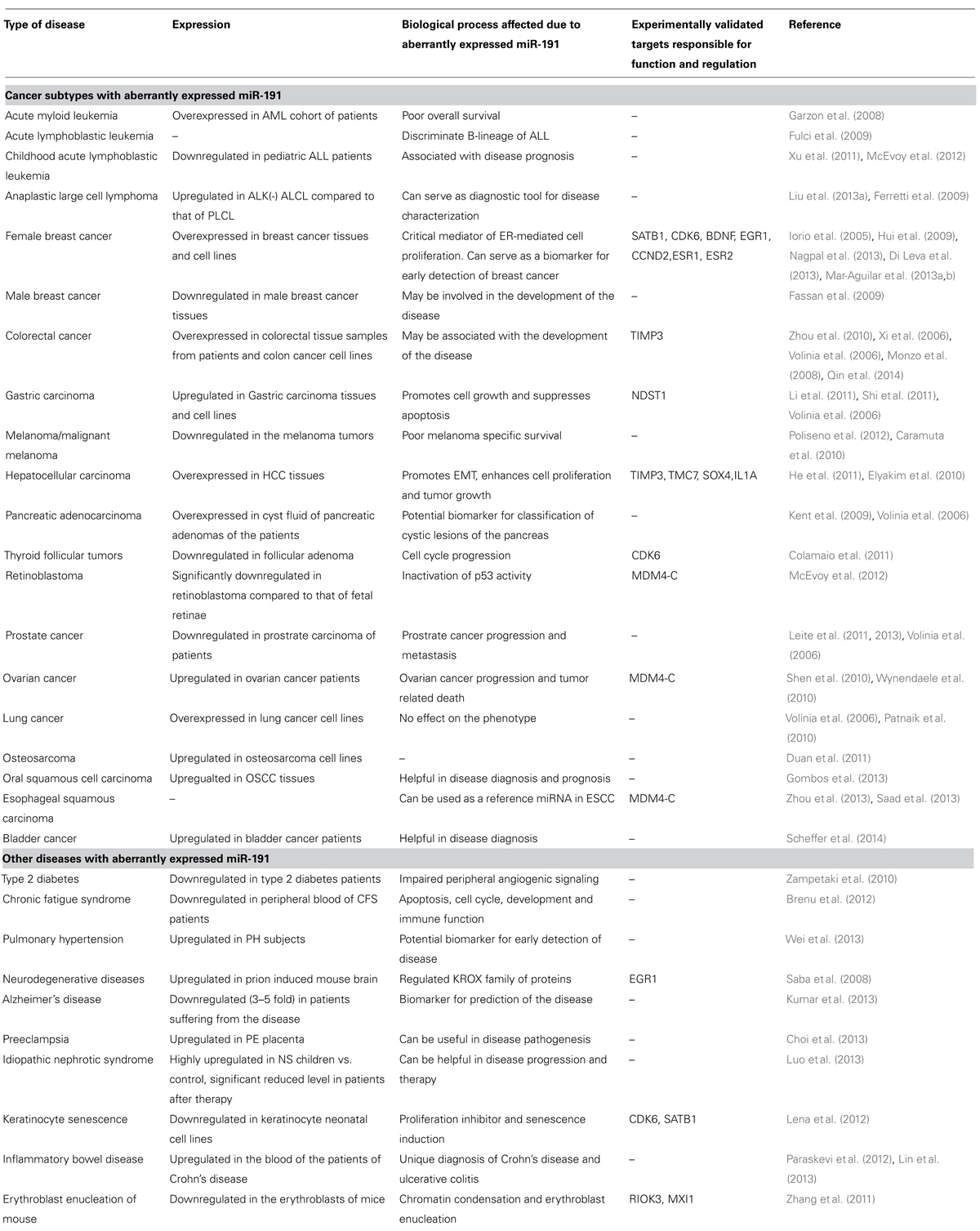
TABLE 1. Summary of disease subtypes reported to have aberrantly expressed miR-191 along with the summary on cellular effects/disease prognosis and experimentally validated target transcripts.
miR-191 as a Biomarker for Cancer Diagnosis and Prognosis
Since miR-191 is one of the highly expressed and stable miRNA in human serum or saliva, it shows potential of being used as non-invasive biomarker in human subjects (Patel et al., 2011). A new method for the disease characterization based upon blood circulating miRNAs was proposed recently. It involved both miR-191 and miR-425 as part of a unique combination of 12 miRNAs that could serve as potential biomarkers to distinguish disease/cancer from healthy controls in 10 out of 13 diseases (p = 0.046; Taguchi and Murakami, 2013). Zhang et al. (2012) recently filed a patent involving a group of 7 miRNAs including miR-191 that serve as markers for prediction, diagnosis, and progression of pancreatic cancer. Similarly, miR-191 (along with miR-182 and miR-199a) based methods for diagnosis and prognosis of AML have been recently patented (Croce, 2013). Higher expression of miR-191 was found to be significantly associated with poor overall survival (OS) and event free survival in adult AML patients (Garzon et al., 2008). Also, in colorectal cancer patients, higher miR-191 expression has been shown to be associated with clinical stage, metastasis and tumor invasion (Xi et al., 2006). In contrast, reduced miR-191 levels have been reported to be associated with bad prognosis or short OS or disease relapse in melanoma, pediatric AML or male smoker squamous cell carcinoma (SCC) patients (Mueller et al., 2009; Zhang et al., 2009; Caramuta et al., 2010; Landi et al., 2010; Xu et al., 2011).
Apart from miR-191 alone as a prognostic marker, the miR-146b/miR-191 ratio has also been reported as a negative prognostic indicator in lung SCC patients (0.29+0.25; Ernest et al., 2012). Likewise, Mar-Aguilar et al. (2013a) have shown that combined expression profile of two miRNAs (miR-125b/miR-191 and miR-21/miR-191) is more specific in discriminating between breast cancer and non-tumor tissue (Mar-Aguilar et al., 2013a). Interestingly, presence of genetic variants in miR-191 precursor has been linked to genetic predisposition to familial ovarian cancer and could therefore serve as a diagnostic marker (Duan et al., 2011). Additionally, epigenetic regulation (hypomethylation) of miR-191 locus has been associated with poor prognosis in hepatocellular carcinoma (HCC; He et al., 2011). Hence, quantification of miR-191/its precursor variants/methylation patterns of its promoter using tissue/blood samples may emerge as a novel method for cancer diagnosis and prognosis.
miR-191 as a Regulator of Various Hallmarks of Cancer
While there are ample reports on abnormal miR-191 levels in various cancers, the knowledge about its functional impact is quite limited. Studies in breast, colon, gastric, and hepatic cancer cell lines suggest miR-191 to be an oncomiR (Xi et al., 2006; Elyakim et al., 2010; Shi et al., 2011; Di Leva et al., 2013; Nagpal et al., 2013). miR-191 was shown to be involved in regulation of cell proliferation, apoptosis, and epithelial mesenchymal transition in HCC (Elyakim et al., 2010; He et al., 2011). In colon cancer, anti-miR-191 was shown to attenuate the invasiveness, suppress proliferation and induce apoptosis and in gastric cancer, miR-191 overexpression was shown to enhance cell proliferation and reduce apoptosis (Shi et al., 2011; Qin et al., 2014). However, functional analysis of miR-191 levels in pancreatic ductal carcinoma cells showed that although it is responsible for maintaining the transformed stage but cell proliferation is not affected (Kent et al., 2009). Recently, Nagpal et al. (2013) showed that miR-191 is responsible for promotion of various hallmarks of cancer (proliferation, metastasis, stress resistance) in ER+ breast cancer (MCF7) cells, while Di Leva et al. (2013) demonstrated that miR-191/425 cluster is responsible for reduced proliferation, tumorigenesis, and metastasis in aggressive ER- breast cancer cells (MDA-MB-231; Di Leva et al., 2013; Nagpal et al., 2013). This dual effect of miR-191 could be due to the fact that miR-191 is an ER regulated miRNA that functions as a critical mediator of estrogen mediated cell proliferation. miR-191 has been shown to regulate cell cycle progression in breast cancer by promoting G1/S and G2/M transitions as the cells overexpressing miR-191 entered S-phase much faster with drastic reduction in G0/G1 phase.
In contrast, miR-191 has been projected as a tumor-suppressor microRNA that leads to reduced growth and migration in thyroid follicular carcinoma (Colamaio et al., 2011). Notably, though miR-191 was shown to be differentially expressed (significantly upregulated in both squamous and non-squamous NSCLC, p < 1e-07) in lung cancer, it was shown to not alter cell cycle, proliferation or chemosensitivity of lung cancer cell lines (Volinia et al., 2006; Yanaihara et al., 2006; Patnaik et al., 2010).
miR-191 as a Therapeutic Target in Cancer
Till date only two groups have performed mouse xenograft assays to test miR-191 activity in vivo evaluating its therapeutic potential and the results are certainly encouraging. Elyakim et al. (2010) generated an orthotopic human liver tumor xenografts in nude mice and administered 2-O-metoxyethyl (MOE) anti-miR-191 through repeated intraperitoneal injections. A statistically significant reduction in tumor mass was observed without toxicity by the end of 40 days (Elyakim et al., 2010). Similarly, a subcutaneous transplantation of the anti-miR-191 transfected ER-alpha positive MCF7 or ZR-75-1 breast cancer cells in nude mice resulted in a 50% reduction in tumor mass. The effect of miR-191 on metastasis was also studied in aggressive breast cancer. Injection of miR-191 overexpressing ER negative breast cancer cells (MDA-MB-231) in NOD-SCID mice led to a significant reduction in a number of micrometastasis to lungs of mice as compared to the control tumor cells (Di Leva et al., 2013). Thus, regulated expression of miR-191 may serve as potential therapeutic treatment of the disease.
Regulation of miR-191 Expression in Cancer
Despite well documented miR-191 and cancer links, very little is known about the factors that lead to its deregulation in various tumor cells. In breast cancer, two independent groups demonstrated that estrogen and tumor microenvironment play a major role in regulation of miR-191 (Di Leva et al., 2013; Nagpal et al., 2013). The promoter of miR-191 bears estrogen response elements which show dynamic binding of ER-alpha and ER-beta transcription factors in response to estrogen treatment (Nagpal et al., 2013). The levels of both miR-191 and miR-425 have been reported to be estrogen inducible (Di Leva et al., 2013; Nagpal et al., 2013). Tumor microenvironment (hypoxia and nutrient deprivation) has also been reported to strongly induce miR-191 in breast cancer (Nagpal et al., 2013). A report by Xi et al. (2006) suggests that the loss of p53 may be the reason behind higher expression (median 1.4-fold, p 0.0264) of miR-191 in colorectal tumor. Another study shows that the gain in expression of MDM4, a miR-191 target gene, in retinoblastoma leads to inactivation of p53 activity (McEvoy et al., 2012). Interestingly, the hypomethylation of CpG islands in miR-191 promoter has been linked to higher expression of miR-191 in HCC (He et al., 2011). While in melanoma, both, the genomic loss of the miR-191 locus and downregulation of miR-191 by a transcription factor, SNAIL have been shown as the reasons for low miR-191 levels (Mueller et al., 2009; Poliseno et al., 2012). To conclude, transcription factors, tumor microenvironment and epigenetic mechanisms play major role in regulation of miR-191 in various cancers.
Several recent reports suggest that miR-191 levels are also responsive to chemical drugs and radiation treatments. Chaudhry et al. (2013) have shown miR-191 induction in response to X-ray treatment in human lymphoblast cells (TK6 cell line). However, miR-191 was shown to be rather downregulated in two colon cancer cell lines HCT116 and HCT-8 when treated with chemotherapeutic drugs (5-Fluorouracil and Oxaliplatin; Zhou et al., 2010). Thus, the effects on miR-191 levels may vary depending on the stress or tissue/cell type. Work in our lab shows that miR-191 overexpressing breast cancer cells show increased survival in response to chemotherapeutic drug or hypoxia treatment (Nagpal et al., 2013). Altogether these reports suggest prospects of miR-191 being a target for studying chemotherapeutic interference (Zhou et al., 2010).
Interestingly, carcinogens too have been reported to mediate miR-191 regulation. Davidson et al. (2009) showed that AOM (carcinogen) treated rats show downregulation of miR-191. Conversely, feeding rats with fish oil rich in polyunsaturated fatty acids (n = 3 PUFA; a chemoprotective agent) prevented AOM mediated downregulation of miR-191 and tumor formation (Davidson et al., 2009). Environmental carcinogens also play a major role in regulation of miR-191, for example, cigarette smoke and TCDD (a dioxin family carcinogen) was shown to regulate miR-191 levels in rat lungs and hepatic cancer, respectively (Izzotti et al., 2009; Elyakim et al., 2010). Thus, these data provide evidence for hormonal, environmental and dietary regulation of miR-191.
miR-191 in Diseases Other than Cancer
Association of miR-191 is not only limited to various cancers but its irregular expression and functional abnormalities have been reported in a variety of other diseases as well (Table 1). The functional impact of miR-191 is still unknown in these diseases but it does show potential as a diagnostic or prognostic marker for some of them. Recently Lin et al. (2013) reported that miR-191 may be a diagnostic biomarker candidate that could help to distinguish between the Crohn’s disease (CD) and ulcerative colitis (UC; Lin et al., 2013). Further, as the two disease types differ in their associated T- and B- cells, miR-191 may be involved in regulation of innate and adaptive immunity as well (Paraskevi et al., 2012). Similarly, miR-191 was shown to be a potential biomarker for early detection and severity of pulmonary hypertension (Wei et al., 2013). Remarkably, miR-191-5p was shown to be one of the best biomarker candidates to predict the Alzheimer’s disease with >95% accuracy (Kumar et al., 2013). miR-191 was also found to be significantly induced (3.79-fold, p < 0.0001) in nephrotic syndrome (NS) children making it a potential diagnostic marker for pediatric NS. Interestingly, NS patients under remission have significantly reduced level of miR-191 suggestive of its association with disease progression and therapy (Luo et al., 2013). Overall, miR-191 seems to be closely associated with the pathogenesis of diverse diseases and may also be involved in innate immune responses. A compiled list of diseases having an association with miR-191 is given in Table 1.
miR-191 Targets
A number of miR-191 targets have been validated and characterized functionally. Its targets range from chromatin remodelers to transcription factors to cell cycle regulators. A detailed analysis of the validated potential targets of miR-191 is given below:
Transcription Factors and Chromatin Regulators
SATB1 (special AT-rich sequence-binding protein-1)
Special AT-rich sequence-binding protein-1 is a global chromatin remodeler and transcription factor that is known to be overexpressed in several cancers and also associated with aggressive phenotype in colorectal and breast cancer (Cai et al., 2006; Han et al., 2008; Meng et al., 2012). miR-191 mediated downregulation of SATB1 has been linked to gain/loss of epithelial/mesenchymal markers in aggressive breast cancer and enhanced cell proliferation and migration in hormone dependent breast cancer (Di Leva et al., 2013; Nagpal et al., 2013). The miR-191/SATB1 functional link has been proposed to be helpful for prognosis and therapeutics of breast cancer (Nagpal et al., 2013). Additionally, suppression of SATB1 was demonstrated to be linked to cell senescence, as its suppression by miR-191 led to enhanced senescence in keratinocytes (Lena et al., 2012).
RIOK3 (RIO kinase 3) and MXI1 (max-interacting protein 1)
RIO kinase 3 and max-interacting protein 1 are reported to be negatively regulated by miR-191 and play essential role in chromatin condensation and enucleation during mouse erythroid differentiation (Zhang et al., 2011).
EGR1 (early growth response protein 1)
Early growth response protein 1 is a zinc finger transcription factor mainly involved in cell growth, development and stem cell homeostasis (Yan et al., 2000). It was demonstrated to be an estrogen responsive miR-191 target, having regulatory role in cell proliferation in ER-alpha positive breast cancer cells. Additionally, it was reported to be downregulated by miR-191 in scrapie infection, a prion disease (Saba et al., 2008; Di Leva et al., 2013).
SOX4 SRY (sex determining region Y)-box 4
Sex determining region Y-box 4 is a transcriptional factor found to be upregulated in various cancers (Rhodes et al., 2004). It is involved in regulation of development, differentiation, proliferation, apoptosis, and more recently epithelial-mesenchymal-transition (Pan et al., 2009). It was shown to be a novel target of miR-191 in HCC (Elyakim et al., 2010). Thus, downregulation of SOX4 by miR-191 may affect chromatin architecture and crucial cellular processes.
MDM4 (Mdm4 p53 binding protein homolog)
Mdm4 p53 binding protein homolog is a negative regulator of transcription factor, p53 and is found to be overexpressed in 17% of all cancers (Liu et al., 2012). miR-191 is reported to regulate cancer progression and chemosensitivity in ovarian cancer by modulating the level of MDM4 through binding with an illegitimate site (rs4245739) within the 3′ UTR (Wynendaele et al., 2010). Recently, the same SNP (rs4245739) was investigated and found to be a risk factor for esophageal SCC and breast cancer (Zhou et al., 2013). The MDM4 rs4245739 SNP alone or in combination with the p53 variant Arg72Pro has been shown to be responsible for attenuating breast cancer risk in Chinese population (Liu et al., 2013b).
Cell Cycle Regulators
CDK6 (cyclin-dependent kinase 6)
Cyclin-dependent kinase 6 is a protein kinase mainly associated with cell cycle progression and differentiation (Robker and Richards, 1998; Depamphilis et al., 2012). It has been reported as a bona fide target of miR-191 in aggressive and hormone receptor positive breast cancer and thyroid and follicular carcinoma (Colamaio et al., 2011; Di Leva et al., 2013; Nagpal et al., 2013). Additionally, miR-191 mediated downregulation of CDK6 has been reported to trigger senescence in keratinocytes (Lena et al., 2012).
CCND2 (cyclin D2)
Cyclin D2 is a cell cycle regulatory protein involved in estrogen mediated cell proliferation (Robker and Richards, 1998). It was recently established as a target of miR-191 and its downregulation led to suppression of PI3/AKT pathway thereby leading to reduced cell proliferation in aggressive breast cancer (Di Leva et al., 2013).
Others
NDST1 (N-deacetylase/N-sulfotransferase1)
N-deacetylase/N-sulfotransferase1 is a key enzyme involved in the biosynthesis of heparan sulfate chains and is responsible for maintenance of physiological functions (Sheng et al., 2011). It regulates binding of FGF to mammary epithelial cells leading to branching morphogenesis (Crawford et al., 2010). It was shown to be negatively regulated by miR-191 and its downregulation has suppressive effect on cell proliferation in gastric carcinoma (Shi et al., 2011).
BDNF (brain-derived neurotrophic factor)
Brain-derived neurotrophic factor is a neurotrophin essential for differentiation, survival, and synaptic plasticity of CNS (Numakawa et al., 2011). Recently our group has shown it to be a target of miR-191 in ER positive breast cancer (Nagpal et al., 2013).
TIMP3 (tissue inhibitor of metalloprotease 3)
Tissue inhibitor of metalloprotease 3 is a proapoptotic protein whose expression is negatively correlated with cell growth and invasiveness (Kashiwagi et al., 2001). It is a direct miR-191 target in colorectal carcinoma and its targeted downregulation is associated with enhanced invasiveness of the disease (Qin et al., 2014).
An overview of miR-191 regulatory components along with its functional impact on direct and indirect targets is given in Figure 1.
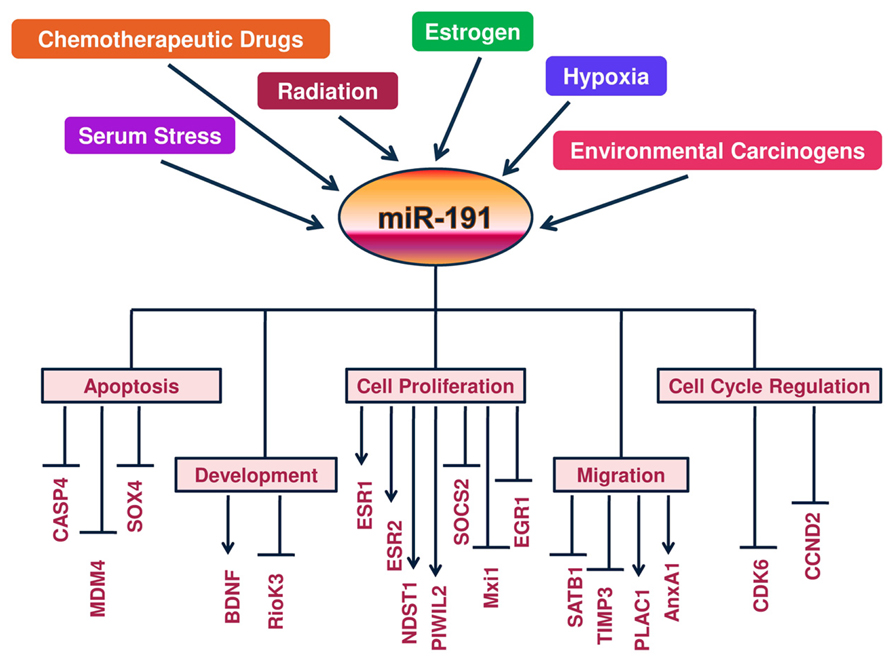
FIGURE 1. A Schematic view of regulation of miR-191 and its targets: a molecular model is presented whereby hormones (estrogen), tumor microenvironment (hypoxia and serum stress) and DNA damage signals (chemodrugs, radiation and carcinogens) regulate miR-191 levels in the cancer cells. In response, miR-191 controls the expression of a number of genes involved in a variety of cellular processes (ranging from cell proliferation to apoptosis to cell migration) to modulate the development of cancer. Overall, miR-191 integrates multiple pathogenic signals to regulate cancer phenotype.
Concluding Remarks and Future Perspectives
Our review brings to limelight an important player in cancer biology that has been shown to have profound effects on various hallmarks of cancer. Due to its ubiquitous role in various biological processes, the interest in miRNA-191 is likely to dramatically increase in coming years, especially in cancer. Though various studies document miR-191 association with cancer progression, but its link to cancer initiation has not yet been elucidated. However, since carcinogens have been shown to regulate the levels of miR-191, it may be involved in tumor initiation process as well.
Considering the widespread association of miR-191 to several cancer types and recent studies documenting that its inhibition leads to reversal of cancer phenotype, it is tempting to speculate that miR-191 may qualify as a candidate for “oncogene addition.” However, the validation of this concept warrants careful and extensive experimentation using genetically engineered mouse models of miR-191. These models may also throw light on the role of miR-191 in various stages of tumor development. Nevertheless, based on miR-191-lung cancer studies, miR-191 may not necessarily functionally contribute to aggression of all cancer/diseases where its levels are abnormally expressed (Patnaik et al., 2010).
Interestingly, several types of abnormalities have been linked to miR-191 in cancer. These include but may not be limited to aberrant mature miR-191 levels, hypomethylated miR-191 promoter, mutations in miR-191 target site or loss of genomic locus (Caramuta et al., 2010; He et al., 2011). However, there are very few reports depicting the basis behind its altered expression in disease states compared to that of normal. Till date transcription factors, tumor microenvironment and epigenetic regulation have been identified as the major players responsible for aberrant expression of miR-191 in various disease states (He et al., 2011; Di Leva et al., 2013; Nagpal et al., 2013). A strong estrogen and hypoxic regulation of miR-191 have been shown to regulate key pathways in breast cancer (Di Leva et al., 2013; Nagpal et al., 2013). In light of recent reports of interdependence of estrogen and hypoxia pathways, it would be interesting to know if miR-191 functions to connect the two pathways. Further, considering miR-191 to be stress inducible and its reported effects on apoptosis, it would be interesting to see if p53 is directly involved in its regulation.
Owing to highly consistent and stable expression of miR-191 and the ability to detect it in human saliva or serum, miR-191 emerges as a strong candidate for non-invasive disease diagnostics (Patel et al., 2011). A strong association between miR-191 levels and disease prognosis/survival laid basis for two recent patents featuring miR-191 based diagnosis and prognosis in AML and pancreatic cancer (Zhang et al., 2012; Croce, 2013). It is likely that miR-191 may emerge as an independent or a co-prognostic marker in other cancers as well. Use of disease specific mouse models, conditional transgenic as well as knockouts are required to elucidate its functional role in vivo and verify its therapeutic potential, both of which remain largely unexplored till now. More novel approaches such as combination of Argonaute protein immunoprecipitation, deep sequencing, or proteomic profiling approaches are required to identify its potential bona fide targets responsible for its functional impact. The fact that miR-191 may directly enhance target transcript levels is striking and needs further elucidation of the mechanism. In particular a combination of phenotypic, transcriptomic, proteomic, and metabolomic changes on manipulation of miR-191 levels in cell line or mouse models will provide a complete understanding of miR-191 functions. To conclude, achieving ways to regulate the level of this particular microRNA in patients (in a safe and consistent manner) might help to fight the underlying disease.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Altuvia, Y., Landgraf, P., Lithwick, G., Elefant, N., Pfeffer, S., Aravin, A., et al. (2005). Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 33, 2697–2706. doi: 10.1093/nar/gki567
Bartel, D. P., and Chen, C. Z. (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5, 396–400. doi: 10.1038/nrg1328
Brenu, E. W., Ashton, K. J., van Driel, M., Staines, D. R., Peterson, D., Atkinson, G. M., et al. (2012). Cytotoxic lymphocyte microRNAs as prospective biomarkers for Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J. Affect. Disord. 141, 261–269. doi: 10.1016/j.jad.2012.03.037
Cai, S., Lee, C. C., and Kohwi-Shigematsu, T. (2006). SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288. doi: 10.1038/ng1913
Caramuta, S., Egyházi, S., Rodolfo, M., Witten, D., Hansson, J., Larsson, C., et al. (2010). MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J. Invest. Dermatol. 130, 2062–2070. doi: 10.1038/jid.2010.63
Cekaite, L., Clancy, T., and Sioud, M. (2010). Increased miR-21 expression during human monocyte differentiation into DCs. Front. Biosci. (Elite Ed.) 2:818–828. doi: 10.2741/E143
Chaudhry, M. A., Omaruddin, R. A., Brumbaugh, C. D., Tariq, M. A., and Pourmand, N. (2013). Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J. Radiat. Res. 54, 808–822. doi: 10.1093/jrr/rrt014
Choi, S. Y., Yun, J., Lee, O. J., Han, H. S., Yeo, M. K., Lee, M. A., et al. (2013). MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta 34, 799–804. doi: 10.1016/j.placenta.2013.06.006
Colamaio, M., Borbone, E., Russo, L., Bianco, M., Federico, A., Califano, D., et al. (2011). miR-191 down-regulation plays a role in thyroid follicular tumors through CDK6 targeting. J. Clin. Endocrinol. Metab. 96, E1915–E1924. doi: 10.1210/jc.2011-0408
Crawford, B. E., Garner, O. B., Bishop, J. R., Zhang, D. Y., Bush, K. T., Nigam, S. K., et al. (2010). Loss of the heparan sulfate sulfotransferase, Ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS ONE 5:e10691. doi: 10.1371/journal.pone.0010691
Croce, C. M. (2013). MiR-182-, miR-191, miR-199a-based methods for the diagnosis and prognosis of acute myeloid leukemia (AML), US 8431342.
Davidson, L. A., Wang, N., Shah, M. S., Lupton, J. R., Ivanov, I., and Chapkin, R. S. (2009). n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis 30, 2077–2084. doi: 10.1093/carcin/bgp245
Depamphilis, M. L., de Renty, C. M., Ullah, Z., and Lee, C. Y. (2012). “The octet”: eight protein kinases that control mammalian DNA replication. Front. Physiol. 3:368. doi: 10.3389/fphys.2012.00368
Di Leva, G., Piovan, C., Gasparini, P., Ngankeu, A., Taccioli, C., Briskin, D., et al. (2013). Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 9:e1003311. doi: 10.1371/journal.pgen.1003311
Duan, Z., Choy, E., Harmon, D., Liu, X., Susa, M., Mankin, H., et al. (2011). MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 10, 1337–1345. doi: 10.1158/1535-7163.MCT-11-0096
Elyakim, E., Sitbon, E., Faerman, A., Tabak, S., Montia, E., Belanis, L., et al. (2010). hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 70, 8077–8087. doi: 10.1158/0008-5472.CAN-10-1313
Ernest, N., Chen, G., Chang, A. C., Lin, J., Reddy, R., Orringer, M. B., et al. (2012). Ratio of miR-146b/miR-191 in serum predicts prognosis in surgically resected lung squamous cell carcinomas. Cancer Res. 72:4147. doi: 10.1158/1538-7445.AM2012-4147
Fassan, M., Baffa, R., Palazzo, J. P., Lloyd, J., Crosariol, M., Liu, C. G., et al. (2009). MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 11:R58. doi: 10.1186/bcr2348
Ferretti, E., De Smaele, E., Po, A., Di Marcotullio, L., Tosi, E., Espinola, M. S., et al. (2009). MicroRNA profiling in human medulloblastoma. Int. J. Cancer 124, 568–577. doi: 10.1002/ijc.23948
Fulci, V., Colombo, T., Chiaretti, S., Messina, M., Citarella, F., Tavolaro, S., et al. (2009). Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer 48, 1069–1082. doi: 10.1002/gcc.20709
Garzon, R., Volinia, S., Liu, C. G., Fernandez-Cymering, C., Palumbo, T., Pichiorri, F., et al. (2008). MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111, 3183–3189. doi: 10.1182/blood-2007-07-098749
Gombos, K., Horváth, R., Szele, E., Juhász, K., Gocze, K., Somlai, K., et al. (2013). miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res. 33, 1511–1517.
Griffiths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A., and Enright, A. J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144. doi: 10.1093/nar/gkj112
Han, H. J., Russo, J., Kohwi, Y., and Kohwi-Shigematsu, T. (2008). SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452, 187–193. doi: 10.1038/nature06781
He, Y., Cui, Y., Wang, W., Gu, J., Guo, S., Ma, K., et al. (2011). Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia 13, 841–853.
Hui, A. B., Shi, W., Boutros, P. C., Miller, N., Pintilie, M., Fyles, T., et al. (2009). Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab. Invest. 89, 597–606. doi: 10.1038/labinvest.2009.12
Inoue, T., Iinuma, H., Ogawa, E., Inaba, T., and Fukushima, R. (2012). Clinicopathological and prognostic significance of microRNA-107 and its relationship to DICER1 mRNA expression in gastric cancer. Oncol. Rep. 27, 1759–1764. doi: 10.3892/or.2012.1709
Iorio, M. V., Ferracin, M., Liu, C. G., Veronese, A., Spizzo, R., Sabbioni, S., et al. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070. doi: 10.1158/0008-5472.CAN-05-1783
Izzotti, A., Calin, G. A., Arrigo, P., Steele, V. E., Croce, C. M., and De Flora, S. (2009). Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 23, 806–812. doi: 10.1096/fj.08-121384
Kasashima, K., Nakamura, Y., and Kozu, T. (2004). Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 322, 403–410. doi: 10.1016/j.bbrc.2004.07.130
Kashiwagi, M., Tortorella, M., Nagase, H., and Brew, K. (2001). TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5). J. Biol. Chem. 276, 12501–12504. doi: 10.1074/jbc.C000848200
Kent, O. A., Mullendore, M., Wentzel, E. A., López-Romero, P., Tan, A. C., Alvarez, H., et al. (2009). A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol. Ther. 8, 2013–2024. doi: 10.4161/cbt.8.21.9685
Kiezun, A., Artzi, S., Modai, S., Volk, N., Isakov, O., and Shomron, N. (2012). miRviewer: a multispecies microRNA homologous viewer. BMC Res. Notes 5:92. doi: 10.1186/1756-0500-5-92
Kim, J., Krichevsky, A., Grad, Y., Hayes, G. D., Kosik, K. S., Church, G. M., et al. (2004). Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 101, 360–365. doi: 10.1073/pnas.2333854100
Kumar, P., Dezso, Z., MacKenzie, C., Oestreicher, J., Agoulnik, S., Byrne, M., et al. (2013). Circulating miRNA biomarkers for Alzheimer’s disease. PLoS ONE 8:e69807. doi: 10.1371/journal.pone.0069807
Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. (2003). New microRNAs from mouse and human. RNA 9, 175–179. doi: 10.1261/rna.2146903
Landi, M. T., Chatterjee, N., Caporaso, N. E., Rotunno, M., Albanes, D., Thun, M., et al. (2010). GPC5 rs2352028 variant and risk of lung cancer in never smokers. Lancet Oncol. 11, 714–716. doi: 10.1016/S1470-2045(10)70096-6
Leite, K. R., Tomiyama, A., Reis, S. T., Sousa-Canavez, J. M., Sañudo, A., Camara-Lopes, L. H., et al. (2013). MicroRNA expression profiles in the progression of prostate cancer-from high-grade prostate intraepithelial neoplasia to metastasis. Urol. Oncol. 31, 796–801. doi: 10.1016/j.urolonc.2011.07.002
Leite, K. R., Tomiyama, A., Reis, S. T., Sousa-Canavez, J. M., Sañudo, A., Dall’Oglio, M. F., et al. (2011). MicroRNA-100 expression is independently related to biochemical recurrence of prostate cancer. J. Urol. 185, 1118–1122. doi: 10.1016/j.juro.2010.10.035
Lena, A. M., Mancini, M., Rivetti di Val Cervo, P., Saintigny, G., Mahé, C., Melino, G., et al. (2012). MicroRNA-191 triggers keratinocytes senescence by SATB1 and CDK6 downregulation. Biochem. Biophys. Res. Commun. 423, 509–514. doi: 10.1016/j.bbrc.2012.05.153
Li, X., Zhang, Y., Zhang, H., Liu, X., Gong, T., Li, M., et al. (2011). miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 9, 824–833. doi: 10.1158/1541-7786.MCR-10-0529
Lin, J., Cao, Q., Zhang, J., Li, Y., Shen, B., Zhao, Z., et al. (2013). MicroRNA expression patterns in indeterminate inflammatory bowel disease. Mod. Pathol. 26, 148–154. doi: 10.1038/modpathol.2012.131
Liu, C., Iqbal, J., Teruya-Feldstein, J., Shen, Y., Dabrowska, M. J., Dybkaer, K., et al. (2013a). MicroRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood 122, 2083–2092. doi: 10.1182/blood-2012-08-447375
Liu, J., Tang, X., Li, M., Lu, C., Shi, J., Zhou, L., et al. (2013b). Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res. Treat. 140, 151–157. doi: 10.1007/s10549-013-2615-x
Liu, L., Fan, L., Fang, C., Zou, Z. J., Yang, S., Zhang, L. N., et al. (2012). S-MDM4 mRNA overexpression indicates a poor prognosis and marks a potential therapeutic target in chronic lymphocytic leukemia. Cancer Sci. 103, 2056–2063. doi: 10.1111/cas.12008
Luo, Y., Wang, C., Chen, X., Zhong, T., Cai, X., Chen, S., et al. (2013). Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin. Chem. 59, 658–666. doi: 10.1373/clinchem.2012.195297
Mar-Aguilar, F., Luna-Aguirre, C. M., Moreno-Rocha, J. C., Araiza-Chávez, J., Trevino, V., Rodriquez-Padilla, C., et al. (2013a). Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac. J. Clin. Oncol. 9, 53–59. doi: 10.1111/j.1743-7563.2012.01548.x
Mar-Aguilar, F., Mendoza-Ramírez, J. A., Malagón-Santiago, I., Espino-Silva, P. K., Santuario-Facio, S. K., Ruiz-Flores, P., et al. (2013b). Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers 34, 163–169. doi: 10.1155/2013/259454
McEvoy, J., Ulyanov, A., Brennan, R., Wu, G., Pounds, S., Zhang, J., et al. (2012). Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS ONE 7:e42739. doi: 10.1371/journal.pone.0042739
Mellios, N., Huang, H. S., Grigorenko, A., Rogaev, E., and Akbarian, S. (2008). A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 17, 3030–3042. doi: 10.1093/hmg/ddn201
Meng, W. J., Yan, H., Zhou, B., Zhang, W., Kong, X. H., Wang, R., et al. (2012). Correlation of SATB1 overexpression with the progression of human rectal cancer. Int. J. Colorectal Dis. 27, 143–150. doi: 10.1007/s00384-011-1302-9
Monzo, M., Navarro, A., Bandres, E., Artells, R., Moreno, I., Gel, B., et al. (2008). Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 18, 823–833. doi: 10.1038/cr.2008.81
Mueller, D. W., Rehli, M., and Bosserhoff, A. K. (2009). miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Invest. Dermatol. 129, 1740–1751. doi: 10.1038/jid.2008.452
Na, Y. J., Sung, J. H., Lee, S. C., Lee, Y. J., Choi, Y. J., Park, W. Y., et al. (2009). Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp. Mol. Med. 41, 638–647. doi: 10.3858/emm.2009.41.9.070
Nagpal, N., Ahmad, H. M., Molparia, B., and Kulshreshtha, R. (2013). MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 34, 1889–1899. doi: 10.1093/carcin/bgt107
Numakawa, T., Matsumoto, T., Numakawa, Y., Richards, M., Yamawaki, S., and Kunugi, H. (2011). Protective action of neurotrophic factors and estrogen against oxidative stress-mediated neurodegeneration. J. Toxicol. 2011:405194. doi: 10.1155/2011/405194
Palmieri, A., Pezzetti, F., Brunelli, G., Martinelli, M., Lo Muzio, L., Scarano, A., et al. (2010). Anorganic bovine bone (Bio-Oss) regulates miRNA of osteoblast-like cells. Int. J. Periodontics Restorative Dent. 30, 83–87.
Pan, X., Zhao, J., Zhang, W. N., Li, H. Y., Mu, R., Zhou, T., et al. (2009). Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc. Natl. Acad. Sci. U.S.A. 106, 3788–3793. doi: 10.1073/pnas.0810147106
Paraskevi, A., Theodoropoulosb, G., Papaconstantinouc, I., Mantzarisd, G., Nikitease, N., and Gazoulia, M. (2012). Circulating MicroRNA in inflammatory bowel disease. J. Crohns Colitis 6, 900–904. doi: 10.1016/j.crohns.2012.02.006
Patel, R. S., Jakymiw, A., Yao, B., Pauley, B. A., Carcamo, W. C., Katz, J., et al. (2011). High resolution of microRNA signatures in human whole saliva. Arch. Oral Biol. 56, 1506–1513. doi: 10.1016/j.archoralbio.2011.05.015
Patnaik, S. K., Kannisto, E., and Yendamuri, S. (2010). Overexpression of microRNA miR-30a or miR-191 in A549 lung cancer or BEAS-2B normal lung cell lines does not alter phenotype. PLoS ONE 5:e9219. doi: 10.1371/journal.pone.0009219
Poliseno, L., Haimovic, A., Segura, M. F., Hanniford, D., Christos, P. J., Darvishian, F., et al. (2012). Histology-specific microRNA alterations in melanoma. J. Invest. Dermatol. 132, 1860–1868. doi: 10.1038/jid.2011.451
Qin, S., Zhu, Y., Ai, F., Li, Y., Bai, B., Yao, W., et al. (2014). MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma 61, 27–34. doi: 10.4149/neo_2014_005
Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl. Acad. Sci. U.S.A. 101, 9309–9314. doi: 10.1073/pnas.0401994101
Robker, R. L., and Richards, J. S. (1998). Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol. Endocrinol. 12, 924–940. doi: 10.1210/mend.12.7.0138
Saad, R., Chen, Z., Zhu, S., Jia, P., Zhao, Z., Washington, M. K., et al. (2013). Deciphering the unique microRNA signature in human esophageal adenocarcinoma. PLoS ONE 8:e64463. doi: 10.1371/journal.pone.0064463
Saba, R., Goodman, C. D., Huzarewich, R. L., Robertson, C., and Booth, S. A. (2008). A miRNA signature of prion induced neurodegeneration. PLoS ONE 3:e3652. doi: 10.1371/journal.pone.0003652
Scheffer, A. R., Holdenrieder, S., Kristiansen, G., von Ruecker, A., Müller, S. C., and Ellinger, J. (2014). Circulating microRNAs in serum: novel biomarkers for patients with bladder cancer? World J. Urol. 32, 353–358. doi: 10.1007/s00345-012-1010-2
Shen, J., DiCioccio, R., Odunsi, K., Lele, S. B., and Zhao, H. (2010). Novel genetic variants in miR-191 gene and familial ovarian cancer. BMC Cancer 10:47. doi: 10.1186/1471-2407-10-47
Sheng, J., Liu, R., Xu, Y., and Liu, J. (2011). The dominating role of N-deacetylase/N-sulfotransferase 1 in forming domain structures in heparan sulfate. J. Biol. Chem. 286, 19768–19776. doi: 10.1074/jbc.M111.224311
Shi, X., Su, S., Long, J., Mei, B., and Chen, Y. (2011). MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803. Acta Biochim. Biophys. Sin. (Shanghai) 43, 849–856. doi: 10.1093/abbs/gmr084
Taguchi, Y. H., and Murakami, Y. (2013). Principal component analysis based feature extraction approach to identify circulating microRNA biomarkers. PLoS ONE 8:e66714. doi: 10.1371/journal.pone.0066714
Vaz, C., Ahmad, H. M., Sharma, P., Gupta, R., Kumar, L., Kulshreshtha, R., et al. (2010). Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 11:288. doi: 10.1186/1471-2164-11-288
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261. doi: 10.1073/pnas.0510565103
Wei, C., Henderson, H., Spradley, C., Li, L., Kim, I. K., Kumar, S., et al. (2013). Circulating miRNAs as potential marker for pulmonary hypertension. PLoS ONE 8:e64396. doi: 10.1371/journal.pone.0064396
Wynendaele, J., Böhnke, A., Leucci, E., Nielsen, S. J., Lambertz, I., Hammer, S., et al. (2010). An illegitimate microRNA target site within the 3’ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 70, 9641–9649. doi: 10.1158/0008-5472.CAN-10-0527
Xi, Y., Formentini, A., Chien, M., Weir, D. B., Russo, J. J., Ju, J., et al. (2006). Prognostic values of microRNAs in colorectal cancer. Biomark. Insights 2, 113–121.
Xu, G., Zhang, Y., Jia, H., Li, J., Liu, X., Engelhardt, J. F., et al. (2009). Cloning and identification of microRNAs in bovine alveolar macrophages. Mol. Cell. Biochem. 332, 9–16. doi: 10.1007/s11010-009-0168-4
Xu, L., Liang, Y. N., Luo, X. Q., Liu, X. D., and Guo, H. X. (2011). [Association of miRNAs expression profiles with prognosis and relapse in childhood acute lymphoblastic leukemia]. Zhonghua Xue Ye Xue Za Zhi 32, 178–181.
Yan, S. F., Pinsky, D. J., Mackman, N., and Stern, D. M. (2000). Egr-1: is it always immediate and early? J. Clin. Invest. 105, 553–554. doi: 10.1172/JCI9513
Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M., et al. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198. doi: 10.1016/j.ccr.2006.01.025
Zampetaki, A., Kiechl, S., Drozdov, I., Willeit, P., Mayr, U., Prokopi, M., et al. (2010). Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 107, 810–817. doi: 10.1161/CIRCRESAHA.110.226357
Zhang, C., Liu, R., Wang, C., Ba, Y., Zhang, C., and Zeng, K. (2012). Pancreatic cancer markers, and detecting methods, kits, biochips thereof. EP 2518158.
Zhang, H., Luo, X. Q., Zhang, P., Huang, L. B., Zheng, Y. S., Wu, J., et al. (2009). MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS ONE 4:e7826. doi: 10.1371/journal.pone.0007826
Zhang, L., Flygare, J., Wong, P., Lim, B., and Lodish, H. F. (2011). miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 25, 119–124. doi: 10.1101/gad.1998711
Zhou, J., Zhou, Y., Yin, B., Hao, W., Zhao, L., Ju, W., et al. (2010). 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol. Rep. 23, 121–128. doi: 10.3892/or_00000613
Keywords: miR-191, microRNA, cancer, disease, diagnosis, prognosis
Citation: Nagpal N and Kulshreshtha R (2014) miR-191: an emerging player in disease biology. Front. Genet. 5:99. doi: 10.3389/fgene.2014.00099
Received: 18 February 2014; Accepted: 07 April 2014;
Published online: 23 April 2014.
Edited by:
Lawrence Wing Chi Chan, Hong Kong Polytechnic University, ChinaReviewed by:
Francesca Fanini, Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (S.r.l.) – Istituto di Ricovero e Cura a Carattere Scientifico, ItalyAlan G. Ramsay, King’s College London, UK
Copyright © 2014 Nagpal and Kulshreshtha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ritu Kulshreshtha, RNA-II Lab, Department of Biochemical Engineering and Biotechnology, Indian Institute of Technology Delhi, New Delhi-110016, India e-mail: ritu@dbeb.iitd.ac.in; drritukulshreshtha@gmail.com
 Neha Nagpal
Neha Nagpal Ritu Kulshreshtha
Ritu Kulshreshtha