- 1Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Bandar Sunway, Malaysia
- 2Division of Genetics and Molecular Biology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
As the causative agent of foodborne related illness, Vibrio species causes a huge impact on the public health and management. Vibrio species is often associated with seafood as the latter plays a role as a vehicle to transmit bacterial infections. Hence, antibiotics are used not to promote growth but rather to prevent and treat bacterial infections. The extensive use of antibiotics in the aquaculture industry and environment has led to the emerging of antibiotic resistant strains. This phenomenon has triggered an alarming public health concern due to the increase number of pathogenic Vibrio strains that are resistant to clinically used antibiotics and is found in the environment. Antibiotic resistance and the genes location in the strains can be detected through plasmid curing assay. The results derived from plasmid curing assay is fast, cost effective, sufficient in providing insights, and influence the antibiotic management policies in the aquaculture industry. This presentation aims in discussing and providing insights on various curing agents in Vibrio species. To our best of knowledge, this is a first review written discussing on plasmid curing in Vibrio species.
Introduction
Antibiotic resistant infection has become more challenging to treat with existing antibiotics, leading to infections triggering higher morbidity and mortality, imposing huge costs on our society (Carlet et al., 2011; de Kraker et al., 2011; Finley et al., 2013). This increasing resistance involves many human pathogens including Vibrio species. Vibrio are Gram-negative bacteria possessing a curved rod shape that naturally inhabits the estuarine and marine environment worldwide (Hazen et al., 2010; Letchumanan et al., 2014; Raghunath, 2015). The presence of this bacterium in the marine environment raises human concern on food safety due to the latter potentially causing disease outbreaks depending on the environmental conditions (Ceccarelli et al., 2013). Vibrio cholerae, V. parahaemolyticus, V. vulnificus, V. mimicus, V. fluvialis, and V. alginolyticus are among the species commonly known to cause human illnesses (Theresa and Kumar, 2014).
The occurrence of multidrug-resistant (MDR) bacteria to clinically used antibiotics is a major health issue and a great challenge to the worldwide drug discovery programs (Alanis, 2005). It is well documented that both clinical and environmental Vibrio strains harbors antibiotic resistance traits (Letchumanan et al., 2015; Shrestha et al., 2015; Zavala-Norzagaray et al., 2015). A recent study in Iran has reported multidrug resistance profile towards erythromycin, sulfamethoxazole–trimethoprim and ampicillin in V. cholerae isolated from clinical samples (Tabatabaei and Khorashad, 2015). In India, serogroups O1 of V. cholerae classical biotype and sub serotype, Ogawa was identified among the V. cholerae isolated from clinical strains. All the isolates were reported to be resistant to ampicillin, nalidixic acid, and cotrimoxazole (Shrestha et al., 2015). Besides V. cholerae, V. parahaemolyticus have been isolated both from clinical and environmental samples study in India. A clinical study reported 178 V. parahaemolyticus strains were isolated from 13,607 diarrheal patients admitted in Infectious Diseases Hospital, Kolkata since 2001–2012 (Pazhani et al., 2014). Reyhanath and Kutty (2014) have reported the detection and isolation of multidrug resistant strains of V. parahaemolyticus isolated from a fishing land at South India. In another study, pathogenic and antibiotic resistant V. parahaemolyticus strains and other Vibrio species strains were isolated from seafood in Cochin. Majority of the strains in this study were resistant to ampicillin and multiple drug resistance was prevalent among the isolates (Sudha et al., 2014). In recent years, environmental Vibrio strains have been studied in detail for its potential as a reservoir for the wide spread of antibiotic resistance (Zhang et al., 2012).
Every year, more and more pathogenic Vibrio species have been reported to develop higher resistance towards most of the clinically used antibiotics. Drug resistance is an alarming issue worldwide and is spreading rapidly due to overuse, self-medication or the non-therapeutic use of antimicrobials (Slama et al., 2005). Antibiotics and other chemotherapeutic agents are frequently utilized in aquaculture farms as feed additives or immersion baths to achieve either prophylaxis or therapy (Devi et al., 2009; Manjusha and Sarita, 2011; Rico et al., 2012; Cabello et al., 2013). The excessive usage of antibiotics in agriculture and aquaculture environments has caused the development of multidrug resistance in seafood pathogens such as Vibrio species (Sudha et al., 2014). Usually the emerging of single or multiple antibiotic resistances are closely associated with various antimicrobial used (Manjusha and Sarita, 2011). Tetracycline, quinolone, oxytetracycline, enrofloxacin, sarafloxacin, and florfenicol are among the antibiotics allowed and used in the aquaculture industry to ensure continuous production of seafood (Roque et al., 2001; Yano et al., 2014).
Vibrio species are usually known to be highly susceptible to most clinically used antibiotics (Mala et al., 2014; Shaw et al., 2014; Letchumanan et al., 2015; Zavala-Norzagaray et al., 2015). Most of the genetic determinants that confer resistance to antibiotics are located on plasmids. Acquired antibiotic resistance in bacteria is generally mediated by extrachromosomal plasmids and transferable to other bacteria in the environment through vertical gene transfer or horizontal gene transfer (Manjusha and Sarita, 2011). Horizontal gene transfer is very important in the evolution and transmission of resistance genes between species and includes the transfer of resistance genes from fecal bacteria to environmental bacteria (Baquero et al., 2008). These extrachromosomal DNA sequences may be responsible for the emergence of resistance to multiple antibiotics (Schelz et al., 2006). In recent years, the presence of antibiotic resistance genes detected in Vibrio species have increased and includes β-lactam and penicillin resistance genes penA and blaTEM-1 (Srinivasan et al., 2005; Zhang et al., 2009), chloramphenicol resistance genes catI, catII, catIII, catIV, and floR (Dang et al., 2007, 2008) and tetracycline resistance genes tetA, tetB, tetC, tetD, tetE, tetG, tetH, tetJ, tetY, tetZ (Macauley et al., 2007; Zhang et al., 2009; Kim et al., 2013).
Plasmid-mediated multidrug resistance is one of the most pressing problems in the treatment of infectious diseases. The use of plasmid-curing agents may serve as a possible way to eliminate the plasmid and reduce spreading of antibiotic resistance encoded by antibiotic resistance plasmids (R-plasmids) (Molnar et al., 2003). Plasmid curing occurs naturally through cell division or by treating the cells with any chemical or physical agents (Elias et al., 2013). The inhibition of conjugational transfer of antibiotic resistance plasmid can be used to decrease the spread of antibiotic resistance plasmid in the environment. Inhibition of plasmid replication occurs in various stages and well demonstrated through the “rolling circle” model (replication, partition, and conjugal transfer). This could also be the theoretical basis for the elimination of bacterial virulence in the case of plasmid mediated pathogenicity and antibiotic resistance (Brüssow et al., 2004). The aim of this study is to provide essential insights on the traditional plasmid curing assay in Vibrio.
Plasmid Curing in Vibrio
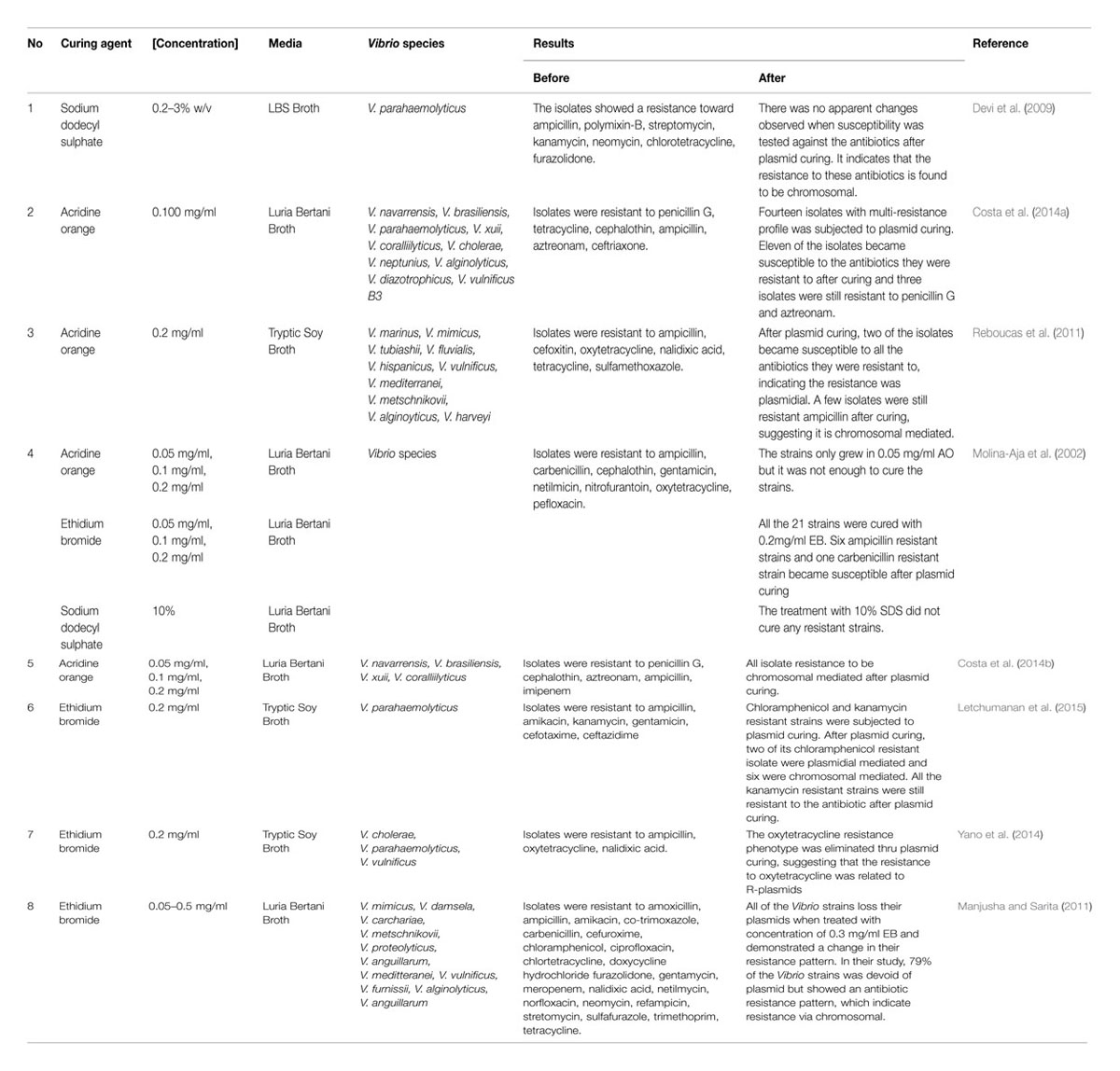
Bacterial plasmids are known to harbor genes for resistances to antibiotics and metals; catabolic pathways such as lactose utilization and degradation of hydrocarbons; and biosynthesis of certain antibiotics. Curing of plasmids from bacteria strains is a way to eliminate the bacteria plasmid and determine the antibiotic resistance mediation. There are several methods involving chemical and physical agents that have been developed to eliminate plasmids. Protocols for plasmid curing in Vibrio consist of chemical agents such as acridine orange (AO), ethidium bromide (EB), and sodium dodecyl sulphate (SDS), and physical agent (Liu et al., 2012) (Table 1). The mechanism of plasmid curing starts from the inhibition of plasmid replication resulted from a single nick, outside of the replication origo of the superhelical structure. The process leads to further relaxation of plasmid DNA, an increase in melting point and circular dichroism. The intercalating agents would then break the superhelical form of plasmid DNA subsequently forming an open circular or linear form plasmid DNA (Spengler et al., 2006). Resistance is usually classified as “chromosomal” when unaffected by plasmid curing and as “plasmidial” when affected.
Intercalating Agents
Intercalating agents such as AO and EB have been successfully used in curing bacterial plasmids. The modes of action of intercalating agents are through preferential inhibition of plasmid replication. Basically, overnight bacteria cultures are inoculated into enrichment broths, Tryptic Soy Broth (TSB), or Luria Bertani Broth (LB). Curing agents at a concentration ranging from 0.1 to 0.5 mg/ml is added to the culture broth. The concentration depends on the organism and curing agent used. The cultures are then incubated overnight at 35 or 37°C under constant agitation. After the treatment, antibiograms assay were again performed to find antibiotic resistance phenotypes.
The effectiveness of a curing agent does vary considerably in ranging of 100- to 1000-fold. This depends on the organisms being treated, curing agent efficiency and efficacy, and the mode of action of respective the curing agent (Carlton and Brown, 1981). Due to these factors, it is essential to use a wide range of curing agent concentration especially when the bacteria are isolated from environmental sources (Trevors, 1986).
Since the 1960s, AO has been used as curing agent and normally involves loss of the whole plasmid (Salisbury et al., 1972; Costa et al., 2014a). In 1970s, Dastidar et al. (1977) reported on the efficiency of AO in eliminating R-plasmids in V. cholerae multidrug-resistant strains. Many other studies have demonstrated the usefulness of AO in clinical strains, animal, or environmental isolates (Kamat and Nair, 1992; Barman et al., 2010; Reboucas et al., 2011; Carvalho et al., 2013; Costa et al., 2014a) although the conventional methods for curing plasmids by curing agents may induce mutations in the host chromosomal DNA (Liu et al., 2012).
Reboucas et al. (2011) have used 0.2 mg/ml AO to cure plasmid of Vibrio species isolated from marine shrimp. The occurrence of multiple antibiotic resistances was observed in 29% (9/31) of Vibrio isolates. Of the total multi-resistant Vibrio isolates submitted to plasmid curing, five (55.5%) lost one or more resistance profile while four isolates (44.5%) did not lose their resistance. Out of the five isolates, two isolates became totally susceptible to all the antibiotics they were resistant to, indicating the resistance was plasmidial. Resistance to oxytetracycline was the most frequently lost phenotype after plasmid curing. The results also demonstrated that isolates were still resistant to ampicillin after plasmid curing, suggesting it could be chromosomal mediated (Reboucas et al., 2011).
Similar results were also demonstrated in another study by Costa et al. (2014a). This study subjected fourteen penicillin G, ampicillin and aztreonam resistant isolates to plasmid curing with 0.1 mg/ml of AO. After the plasmid curing assay, 11 of the isolates resistance changed from resistant to susceptible whereas, three other isolates were still resistant to penicillin G and aztreonam. Teo et al. (2000) noted the ampicillin resistant in Vibrio with a possible via of mediation by β-lactamase blaVHW-1 and blaVHH-1 genes in plasmids. AO has been used in another study by Costa et al. (2014b) and revealed the isolate resistance to be chromosomal mediated after plasmid curing. The loss of phenotype in these studies suggests that AO produce an immediate and complete inhibition of plasmid replication. However, the results may be species dependent and could not be expected with other organisms.
A study was conducted to compare the effectiveness of different plasmid curing agents. Molina-Aja et al. (2002) tested three curing treatments using AO, EB, and SDS. The strains grown in LB was tested with a set of 10% SDS; 0.05, 0.1, and 0.2 mg/ml of AO; 0.05, 0.1, and 0.2 mg/ml of EB. The results stated that the treatment with 10% SDS did not cure any of the study strains. The strains only grew in the lowest concentration of AO (0.05 mg/ml), but it was not enough to cure the stains. All the twenty-one strains were successfully treated with 0.2 mg/ml EB and seven isolates lost their plasmid. All the treated isolates presented changes in their sensitivity toward the antibiotics (Molina-Aja et al., 2002). The authors suggested that EB was selected as a favorable curing agent in comparative to AO because it is difficult to disposal of AO (Molina-Aja et al., 2002).
Ethidium bromide with a formula molecule C21H20N3Br is an intercalating agent which resembles a DNA base pair. Due to its unique structure, EB can easily intercalate into DNA strand. Yano et al. (2014) demonstrated the use of EB to eliminate plasmids in antibiotic resistant Vibrio species isolated from shrimp cultured in inland ponds in Thailand. Typically the resistant isolates were grown in TSB supplemented with 0.2 mg/ml EB. The oxytetracycline resistance phenotype was eliminated through plasmid curing. The authors suggested that the resistance to oxytetracycline was related to R-plasmids (Yano et al., 2014). Our study also utilized 0.2 mg/ml of EB in curing plasmid of V. parahaemolyticus isolates. The study demonstrated chloramphenicol (catA2) gene was detected in eight chloramphenicol resistant V. parahaemolyticus isolates. Two of the isolates had the gene present in their plasmid whereas another six isolates showed possibility of chromosomal-mediated since the isolates exhibit positive amplification with catA2 gene and demonstrated phenotypic resistance to chloramphenicol on the disk diffusion test after plasmid curing. In addition, the result of plasmid curing revealed that kanamycin-resistant V. parahemolyticus were potentially chromosomal mediated since the isolated exhibit positive amplification with aphA-3 gene and demonstrated phenotypic resistance after plasmid curing (Letchumanan et al., 2015).
Manjusha and Sarita (2011) performed plasmid curing using 0.05 to 0.5 mg/ml of EB. It was evident from the curing experiment that all of the Vibrio strains loss their plasmids when treated with concentration of 0.3 mg/ml EB and demonstrated a change in their resistance pattern. In their study, 79% of the Vibrio strains was devoid of plasmid but showed an antibiotic resistance pattern, which indicate chromosomal resistance. The isolates exhibited a chromosomal borne resistance toward amoxicillin, ampicillin, furazolidone, and tetracycline after curing assay. The authors concluded that some of these resistances may be encoded on plasmids in some strains, while the other isolates may be chromosomal mediated.
Sodium Dodecyl Sulfate
Sodium Dodecyl Sulfate is an anionic detergent that is used as a chemical curing agent in Vibrio species. Plasmid containing cells are possibly more sensitive to SDS because of plasmid-specified pili on cell surface. The chemical acts in dislodging the indigenous plasmid from its site of attachment. Curing assay was performed using SDS as the agent in a study done in India. The V. parahaemolyticus isolates were inoculated into LB with different concentrations of SDS (0.2, 0.4, 0.6, 0.8, 1, 2, and 3% w/v). All the strains were still resistant to the antibiotic after curing. The study suggested that the antibiotic resistance of V. parahaemolyticus isolates was chromosomal borne (Devi et al., 2009).
Physical Agents
Based on literature, physical agent such as elevated growth temperature is commonly used in Vibrio plasmid curing. The mode of action of elevated growth temperature is through complete or partial deletions of strain’s plasmid DNA. Elevated incubation temperature (5–7°C) above the optimal growth temperature can be used as a curing method. Study has demonstrated strains that have an optimal growth temperature of 37°C are incubated at 42°C (Carlton and Brown, 1981). The culture are incubated at the elevated temperature until it reaches late log phase, at which time it is diluted (1:20) and re-incubated at the elevated growth temperature until late log phase growth is reached again. A serial dilution is prepared and plated to obtain single colonies which are individually tested for loss of the plasmid-encoded trait and physical absence of the plasmid using agarose gel electrophoresis (Trevors, 1985).
The elevated growth temperature has been successfully used to cure tetracycline resistant, penicillinase-positive strains of Staphylococcus aureus (May et al., 1964). Cultures were grown at 43–44°C to obtain tetracycline-sensitive and penicillinase negative cells. However, these cells did not appear until after several cell generations at the elevated temperature. Elevated incubation temperature (up to 42°C), EB (0.5 mg/ml), 10% SDS, and AO (0.5 mg/ml) were employed to eliminate plasmids from Vibrio species isolated from Mai Po Nature Reserve, Hong Kong (Zhang et al., 2012). The study results stated that none of the plasmid curing agents was effective in eliminating the plasmid from host cells. All attempts to cure the plasmids from their hosts were failed, probably due to the relatively high copy number of the plasmids similar to earlier work (Zhang et al., 2007).
Discussion
Vibrio species occur naturally in the aquatic environments and are normal member of the flora occurring in coastal seawater (Manjusha and Sarita, 2012). In recent years, the increasing number of emerging multi-drug resistant bacteria is distressing. The presence of antibiotic resistant genes in the bacterial plasmid have led to transmission and spreading of drug resistance among pathogenic strains. Several studies have shown conclusively that antibiotic resistance is caused by pressures of clinical antibiotics and use of antibiotics in the agricultural.
Literatures have shown bacterial resistance in Vibrio strains was both plasmid and chromosomal mediated. The studies have demonstrated a high incidence of antibiotic resistance against ampicillin, chloramphenicol, tetracycline, penicillin G, oxytetracycline, carbenicillin, aztreonam, cefuroxime, streptomycin, rifampicin, and amoxicillin. These are among the clinical antibiotics administrated to prevent diseases in human beings. In addition, high numbers of chromosomal mediated antibiotics resistance toward chloramphenicol and ampicillin was observed frequently in the studies. These two antibiotics along with tetracycline, chlortetracycline, nalidixic acid, gentamycin, sulfafurazole, trimethoprim are among the commonly used antibiotics in aquaculture farms through feeds during culture and hatchery production of seeds (Manjusha and Sarita, 2011). The extensive use of these antibiotics in the aquatic environments has caused the Vibrio species to be resistant and carry the resistant genes either in their plasmid or chromosomal. It is noted that plasmid borne integrons are the main players in being able to acquire, rearrange, and express genes conferring antibiotics resistance (Stokes and Hall, 1989; Manjusha and Sarita, 2012). These intergrons have been found in chromosomes of Vibrio species and many other bacteria (Heidelberg et al., 2000; Holmes et al., 2003).
The efficacy of each plasmid curing agent discussed varies depending on the concentration and the organism being cured. Based on the study’s results, it could be concluded that, EB and AO may be a better curing agent than SDS. EB was also preferred by many researchers in comparative to AO because the latter is difficult to be disposed (Molina-Aja et al., 2002). All these chemical curing agents are known to be harmful and cause health problems to human beings. Precaution steps should be followed strictly prior in handling with these curing agent during experiments. When compared with chemical curing agents, physical agent such as elevated growth temperature is least favored in Vibrio plasmid curing studies due to its low successful rate.
Resistance emerges either passively as an aftereffect of pre-existing innate mechanisms or actively through the acquisition of new hereditary material by mobile genetic elements for example plasmids or transposons (Summers, 2006; Wright, 2007). Traditional plasmid curing assay may be used eliminate the bacteria plasmid and detect the antibiotic resistance mediation. This vital information would be beneficial in the global surveillance management of environmental multidrug resistance. Reducing and improving the use of antibiotics in the aquatic environment can reduce resistance and allow the antibiotic to resurface eventually as an effective therapy (Barbosa and Levy, 2000). The establishment of suitable therapeutic doses of antibiotics may also help reduce potential impacts on the environment and on human health (Nogueira-Lima et al., 2006).
In summary, the paper is to provide insight to readers on the traditional plasmid curing agents its effectiveness in Vibrio studies. Nevertheless, a study on Bacillus anthracis has addressed the weakness of chemical and physical plasmid curing agents (Liu et al., 2012). The curing agents are said to cause potential mutation in the host chromosome which interferes with the functional analysis of the plasmid. For this reason, the study developed a curing method using plasmid incompatibility to study Bacillus anthracis plasmid (Liu et al., 2012). But this concern or approach has not been reported in Vibrio studies worldwide. Hence, in view of potential weakness of traditional plasmid curing agents, modern approach to plasmid curing using plasmid incompatibility or next generation sequencing (NGS) could be considered in plasmid curing of Vibrio studies. Another alternative approach would be using microarray technology to detect antibiotic resistant genes in bacteria (Perrenten et al., 2005; Law et al., 2015). To date, a few microarray technologies have been developed for identification of antibiotic resistance genes but are either restricted to a class of drug or limited to a certain number of genes only (Perrenten et al., 2005).
Conclusion
To our knowledge, this is the first presentation that discuss on the traditional plasmid curing in Vibrio species. In the current era of science technology, traditional plasmid curing may be used to eliminate plasmids and determine antibiotic resistance mediation although there is availability of modern methods such as NGS or diagnostic displacement by specific incompatibility. Next generation sequencing has become easier to be accessed, with high throughput results and helps to locate the resistant gene in the genome. However, when compared with traditional plasmid curing, next generation sequencing involves high cost when sequencing genomes of huge samples. Usually, food safety studies involve huge number of samples thus it would be very costly to sequence all the sample isolates genome by using next generation sequencing platform. Hence, alternative approach using traditional plasmid curing is adapted by researchers. The results derived from plasmid curing assay is fast, cost effective, sufficient in providing knowledge and influence the better antibiotic management policies in the aquaculture industry. The aquaculture industry could adapt the method of switching antibiotics used in the aquatic field from time to time in order to allow withdrawal of antibiotic resistance profile in strains. As the effectiveness of clinical antibiotics has declined, the extensive use of antibiotics in the aquaculture and humans are in distress conditions due to horizontal gene transfer and spread of resistant strains. It is very crucial to deal with this threat posed by overused antibiotics in aquaculture promptly.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027) awarded to K-GC. and External Industry Grants from Biotek Abadi Sdn Bhd (vote no. GBA-808138 and GBA-808813) awarded to L-HL.
References
Alanis, A. J. (2005). Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36, 697–705. doi: 10.1016/j.arcmed.2005.06.009
Baquero, F., Martinez, J. L., and Canton, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Barbosa, T. M., and Levy, S. B. (2000). The impact of antibiotic use on resistance development and persistence. Drug Resist. Updat. 3, 303–311. doi: 10.1054/drup.2000.0167
Barman, S., Chatterjee, S., Chowdhury, G., Ramamurthy, T., Niyogi, S. K., Kumar, R., et al. (2010). Plasmid mediated streptomycin and sulfamethoxazole resistance in Shigella flexneri 3a. Inter. J. Antimicrob. Agents. 36, 348–351. doi: 10.1016/j.ijantimicag.2010.06.037
Brüssow, H. H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. J. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Appl. Eviron. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Carlet, J., Collignon, P., Goldmann, D., Goossens, H., Gyssens, I. C., Harbarth, S., et al. (2011). Society’s failure to protect a precious resource: antibiotics. Lancet 378, 369–371. doi: 10.1016/S0140-6736(11)60401-7
Carlton, B. C., and Brown, B. J. (1981). “Gene mutation,” in Manual of Methods for General Bacteriology, eds P. Gerhardt, R. G. E. Murray, R. N. Costiiow, E. W. Nester, W. A. Wood, N. R. Kreig, et al. (Washington, DC: American Society for Microbiology), 222–242.
Carvalho, F. C. T., Sousa, O. V., Carvalho, E. M. R., Hofer, E. and Vieira, R. H. S. F. (2013). Antibiotic resistance of Salmonella spp. isolated from shrimp farming freshwater environment in Northeast region of Brazil. J. Pathol. 2013, 5. doi: 10.1155/2013/685193
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Costa, R. A., Araujo, R. L., Souza, O. V., and Viera, R. H. S. F. (2014a). Antibiotic-resistant vibrios in farmed shrimp. Biomed. Res. Int. 2015, 505914.
Costa, R. A., Colares, L. P., Lima, R. A., Vieira R. H. S. D. F., and de Sousa, S. V. (2014b). Effect of seawater on the activity of antibiotics against vibrios isolated from the hemolymph of cultured pacific white shrimp. J. World Aquac. Soc. 43, 727–732. doi: 10.1111/j.1749-7345.2012.00590.x
Dang, H., Ren, J., Song, L., Sun, S., and An, L. (2008). Dominant chloramphenicol-resistant bacteria and resistance genes in coastal marine waters of Jiaozhou Bay, China. World J. Microbiol. Biotechnol. 24, 209–217. doi: 10.1007/s11274-007-9458-8
Dang, H., Zhang, X., Song, L., Chang, Y., and Yang, G. (2007). Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J. Appl. Microbiol. 103, 2580–2592. doi: 10.1111/j.1365-2672.2007.03494.x
Dastidar, S. G., Poddar, R., Kumar, R., and Chakrabarty, A. N. (1977). Incidence and elimination of R plasmids in Vibrio cholera. Antimicrob. Agents Chemother. 11, 1079–1080. doi: 10.1128/AAC.11.6.1079
de Kraker, M. E., Davey, P. G., Grundmann, H., and BURDEN study group. (2011). Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med. 8:1001104. doi: 10.1371/journal.pmed.1001104
Devi, R., Surendran, P. K., and Chakraborty, K. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the coast of India. World J. Microbiol. Biotech. 25, 2005–2012. doi: 10.1007/s11274-009-0101-8
Elias, H. M., Qader, M. K., and Salih, W. M. (2013). Determination of plasmid DNA role in multidrug resistant Pseudomonas aeruginosa clinical isolates. Inter. J. Microb. Immunol. Res. 1, 80–86.
Finley, R. L., Collignon, P., Larsson, J. D. G., McEwen, S. A., Li, X.-Z., Gaze, W. H., et al. (2013). The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57, 704–710. doi: 10.1093/cid/cit355
Hazen, T. H., Pan, L., Gu, J.-D., and Sobecky, P. A. (2010). The contribution of mobile genetic elements to the evolution and ecology of vibrios. FEMS Microbiol. Ecol. 74, 485–499. doi: 10.1111/j.1574-6941.2010.00937.x
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Holmes, A. J., Gillings, M. R., Nield, B. S., Mabbutt, B. C., Nevalainen, K. M. H., and Stokes, H. W. (2003). The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 5, 383–394. doi: 10.1046/j.1462-2920.2003.00429.x
Kamat, A. S., and Nair, C. K. K. (1992). Evidence for plasmid-mediated toxin production in Bacillus cereus BIS-59. World J. Microbiol. Biotech. 8, 210–211. doi: 10.1007/BF01195851
Kim, M., Kwon, T. H., Jung, S. M., Cho, S. H., Jin, S. Y., Park, N. H., et al. (2013). Antibiotic resistance of bacteria isolated from the internal organs of edible snow crabs. PLoS ONE 8:70887. doi: 10.1371/journal.pone.0070887
Law, J. W., Ab Mutalib, N.-S., Chan, K.-G., and Lee, L.-H. (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Liu, X., Wang, D., Wang, H., Feng, E., and Zhu, L. (2012). Curing of plasmid pXO1 from Bacillus anthracis using plasmid incompatibility. PLoS ONE 7:e29875. doi: 10.1371/journal.pone.0029875
Macauley, J. J., Adams, C. D., and Mormile, M. R. (2007). Diversity of tet resistance genes in tetracycline resistant bacteria isolated from a swine lagoon with low antibiotic impact. Can. J. Microbiol. 53, 1307–1315. doi: 10.1139/W07-104
Mala, E., Oberoi, A., and Alexander, V. S. (2014). Vibrio isolates from cases of acute diarrhea and their antimicrobial susceptibility pattern in a tertiary care hospital. Int. J. Basic Appl. Sci. 3, 35–37.
Manjusha, S., and Sarita, G. B. (2011). Plasmid associated antibiotic resistance in vibrios isolated from coastal waters of Kerala. Inter. Food Res. J. 18, 1171–1181.
Manjusha, S., and Sarita, G. B. (2012). Characterization of plasmids from multiple antibiotic resistant Vibrio sp. isolated from mollucsc and crustaceans. Kor. J. Microbiol. Biotechnol. 40, 197–207. doi: 10.4014/kjmb.1205.05002
May, J. M., Houghton, R. H., and Perret, C. J. (1964). The effect of growth at elevated temperature of some heritable properties of S. aureus. J. Microbiol. 37, 157–169. doi: 10.1099/00221287-37-2-157
Molina-Aja, A., García-Gasca, A., Abreu-Grobois, A., Bolán-Mejía, C., Roque, A., and Gomez-Gil, B. (2002). Plasmid profiling and antibiotic resistance of Vibrio strains isolated from cultured Penaeid shrimp. FEMS Microbiol. Lett. 213, 7–12. doi: 10.1111/j.1574-6968.2002.tb11278.x
Molnar, A., Amaral, L., and Molnar, J. (2003). Antiplasmid effect of promethazine in mixed bacterial cultures. Int. J. Antimicrob. Agents 22, 217–222. doi: 10.1016/S0924-8579(03)00206-1
Nogueira-Lima, A. C., Gesteira, T. C. V., and Mafezoli, J. (2006). Oxytetracycline residues in cultivated marine shrimp (Litopenaeus vannamei Boone, 1931) (Crustacea, Decapoda) submitted to antibiotic treatment. Aquaculture 254, 748–757. doi: 10.1016/j.aquaculture.2005.11.021
Pazhani, G. P., Bhowmik, S. K., Ghosh, S., Guin, S., Dutta, S., Rajendran, K., et al. (2014). Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus Isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 8:e2815. doi: 10.1371/journal.pntd.0002815
Perrenten, V., Vorlet-Fawer, L., Slickers, P., Ehricht, R., Kuhnert, P., and Frey, J. (2005). Microarray-based detection of 90 antibiotic resistance genes of Gram-positive bacteria. J. Clin. Microb. 43, 2291–2302. doi: 10.1128/JCM.43.5.2291-2302.2005
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Reboucas, R. H., Viana de Sousa, O., Sousa Lima, A., Roger F. V., Carvalho, P. B., and Fernandes R. H. V. (2011). Antimicrobial resistance profile of Vibrio species isolated from marine shrimp farming environments (Litopenaeus vannamei) at Cear’a, Brazil. Environ. Res. 111, 21–24. doi: 10.1016/j.envres.2010.09.012
Reyhanath, P. V., and Kutty, R. (2014). Incidence of multidrug resistant Vibrio parahaemolyticus isolated from Ponnani, South India. Iran. J. Microbiol. 6, 60–67.
Rico, A., Satapornvanit, K., Haque, M. M., Min, J., Nguyen, P. T., Telfer, T., et al. (2012). Use of chemicals and biological products in Asian aquaculture and their potential environmental risks: a critical review. Rev. Aqua. 4, 75–93. doi: 10.1111/j.1753-5131.2012.01062.x
Roque, A., Molina-Aja, A., Bolán-Mejía, C., and Gomez-Gil, B. (2001). In vitro susceptibility to 15 antibiotics of vibrios isolated from Penaeid shrimps in Northwestern Mexico. Int. J. Antimicrob. Agents 17, 383–387. doi: 10.1016/S0924-8579(01)00308-9
Salisbury, V., Hedges, R. W., and Datta, N. (1972). Two modes of ‘curing’ transmissible plasmids. J. Gen. Microbiol. 70, 443–452. doi: 10.1099/00221287-70-3-443
Schelz, Z., Molnar, J., and Hohmann, J. (2006). Antimicrobial and antiplasmid activities of essential oils. Fitoterapia 77, 279–285. doi: 10.1016/j.fitote.2006.03.013
Shaw, K. S., Rosenberg Goldstein, R. E., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Cheaspeake Bay and Maryland coastal bay. PLoS ONE 9:e89616. doi: 10.1371/journal.pone.0089616
Shrestha, U. T., Adhikari, N., Maharjan, R., Banjara, M. R., Rijal, K. R., Basnyat, S. R., et al. (2015). Multidrug resistant Vibrio cholerae 01 from clinical and environmental samples in Kathmandu city. BMC Infectious 15:844. doi: 10.1186/s12879-015-0844-9
Slama, T. G., Amin, A., and Brunton, S. A. (2005). A clinician’s guide to the appropriate and accurate use of antibiotics: the Council for Appropriate and Rational Antibiotic Therapy (CARAT) criteria. Am. J. Med. 118, 1–6. doi: 10.1016/j.amjmed.2005.05.007
Spengler, G., Molnar, A., Schelz, Z., Amaral, L., Sharples, D., and Molnar, J. (2006). The mechanism of plasmid curing in bacteria. Curr. Drug. Targets 7, 1–19. doi: 10.2174/138945006777709601
Srinivasan, V., Nam, H. M., Nguyen, L. T., Tamilselvam, B., Murinda, S. E., and Oliver, S. P. (2005). Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog. Dis. 2, 201–211. doi: 10.1089/fpd.2005.2.201
Stokes, H. W., and Hall, R. M. (1989). A novel family of potentially mobile DNA elements encoding site specific gene integration functions: integrons. Mol. Microbiol. 3, 1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x
Sudha, S., Mridula, C., Silvester, R., and Hatha, A. A. M. (2014). Prevalence and antibiotic resistance of pathogenic vibrios in shellfishes from Cochin market. Indian J. Mar. Sci. 43, 815–824.
Summers, A. O. (2006). Genetic linkage and horizontal gene transfer, the roots of the antibiotic multi-resistance problem. Anim. Biotechnol. 17, 125–135. doi: 10.1080/10495390600957217
Tabatabaei, S. M., and Khorashad, A. S. (2015). Antimicrobial resistance patterns of Vibrio cholerae strains isolated from Afgan and Iranian patients in Iran. Int. J. Infect. 2, 22822.
Teo, J. W. P., Suwanto, A., and Poh, C. L. (2000). Novel β-lactamase genes from two environmental isolates of Vibrio harveyi. Antimicrob. Agents Chemother. 44, 1309–1314. doi: 10.1128/AAC.44.5.1309-1314.2000
Theresa, S., and Kumar, P. S. (2014). Isolation, identification and antibiotic resistance pattern of Vibrio species from marine prawns. Inter. J. Biosci. Res. 3, 1–8.
Trevors, J. T. (1985). Bacterial plasmid isolation and purification. J. Microbiol. Methods 3, 259–271. doi: 10.1016/0167-7012(85)90008-9
Trevors, J. T. (1986). Plasmid curing in bacteria. FEMS Microbiol. Rev. 32, 149–157. doi: 10.1111/j.1574-6968.1986.tb01189.x
Wright, G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186. doi: 10.1038/nrmicro1614
Yano, Y., Hamano, K., Satomi, M., Tsutsui, I., Ban, M., and Aue-Umneoy, D. (2014). Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food Cont. 38, 30–45. doi: 10.1016/j.foodcont.2013.09.019
Zavala-Norzagaray, A., Aguirre, A., Velazquez-roman, J., Floresvillaseñor, H., León-sicairos, N., Ley-quiñonez, C., et al. (2015). Isolation, characterization, and antibiotic resistance of Vibrio spp. in Sea Turtles from Northwestern Mexico. Front. Microbiol. 6:635. doi: 10.3389/fmicb.2015.00635
Zhang, R., Pan, L., and Zhao, Z. (2012). High incidence of plasmids in marine Vibrio species isolated from Mai Po Nature Reserve of Hong Kong. Ecotoxicology 21, 1661–1668. doi: 10.1007/s10646-012-0939-7
Zhang, R., Wang, Y., Leung, P. C., and Gu, J.-D. (2007). pVC, a small cryptic plasmid from the environmental isolate of Vibrio cholerae MP-1. J. Microbiol. 45, 193–198
Keywords: Vibrio species, antibiotic, resistance, plasmids, foodborne pathogens
Citation: Letchumanan V, Chan K-G and Lee L-H (2015) An insight of traditional plasmid curing in Vibrio species. Front. Microbiol. 6:735. doi: 10.3389/fmicb.2015.00735
Received: 27 March 2015; Accepted: 06 July 2015;
Published: 17 July 2015.
Edited by:
Pendru Raghunath, Dr. VRK Women’s Medical College, IndiaReviewed by:
Weili Liang, National Institute for Communicable Disease Control and Prevention – Chinese Center for Disease Control and Prevention, ChinaAdrian Canizalez-Roman, Autonomous University of Sinaloa, Mexico
Copyright © 2015 Letchumanan, Chan and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Learn-Han Lee, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Jalan Lagoon Selatan, 46150 Bandar Sunway, Selangor Darul Ehsan, Malaysia, lee.learn.han@monash.edu; leelearnhan@yahoo.com
 Vengadesh Letchumanan
Vengadesh Letchumanan Kok-Gan Chan
Kok-Gan Chan Learn-Han Lee
Learn-Han Lee