- 1The State Key Laboratory of Microbial Technology, School of Life Sciences, Shandong University, Jinan, China
- 2The State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai, China
- 3Shandong Medicinal Biotechnology Center, Shandong Academy of Medical Sciences, Jinan, China
The developmental life cycle of Streptomyces species includes aerial hyphae formation and spore maturation, two distinct developmental processes that are controlled, respectively, by two families of developmental regulatory genes, bld and whi. In this study, we show that the response regulator MtrA (SCO3013) is critical for normal development of aerial hyphae in S. coelicolor and related species. ΔmtrA, a deletion mutant of the response regulator gene mtrA, exhibited the bald phenotype typical of bld mutants defective in aerial mycelium formation, with formation either much delayed or absent depending on the culture medium. Transcriptional analysis indicated that MtrA activates multiple genes involved in formation of aerial mycelium, including chp, rdl, and ram genes, as well as developmental regulatory genes of the bld and whi families. However, the major regulatory gene bldD showed enhanced expression in ΔmtrA, suggesting it is repressed by MtrA. electrophoretic mobility shift assays indicated that MtrA binds upstream of several genes with altered expression in ΔmtrA, including bldD and whiI, and sequences similar to the consensus binding sequence for MtrA of another actinomycete, Mycobacterium tuberculosis, were found in the bound sites. A loosely conserved recognition sequence containing two short, direct repeats was identified for MtrA of S. coelicolor and was validated using mutational analysis. MtrA homologs are widely distributed among Streptomyces species, and as with S. coelicolor, deletion of the mtrA homologs sve_2757 from S. venezuelae and sli_3357 from S. lividans resulted in conditional bald morphology. Our study suggests a critical and conserved role for MtrA in Streptomyces development.
Introduction
Streptomycetes are multicellular, filamentous, Gram-positive bacteria that possess two extraordinary traits rarely seen in other prokaryotes. First, Streptomyces produce a great variety of secondary metabolites that account for more than half of clinically important agents, including antimicrobial and anti-tumor medicines (Hopwood, 2007). Secondly, Streptomyces exhibit a complex developmental life cycle, with the formation of substrate (vegetative) mycelium, aerial mycelium, and spores at different growth stages during development (Flardh and Buttner, 2009; Chater, 2011; McCormick and Flardh, 2011).
Development processes of Streptomyces have been studied primarily in the model strain S. coelicolor (Flardh and Buttner, 2009; Chater, 2011; McCormick and Flardh, 2011). To grow upward into the air, the aerial hyphae need to be coated with a hydrophobic sheath, which is composed mainly of two types of hydrophobic proteins, chaplins and rodlins, whose expression is developmentally regulated (Claessen et al., 2003; Elliot et al., 2003; Talbot, 2003). The eight chaplin proteins of S. coelicolor, ChpA-H, have been categorized into two types according to size and structure. ChpA-ChpC are large proteins with two chaplin domains and a C-terminal sorting signal, whereas ChpD-ChpH are relatively small proteins with only one chaplin domain (Claessen et al., 2003; Elliot et al., 2003). The long chaplin proteins become covalently attached to the cell wall peptidoglycan of the aerial hyphae and spores by sortase enzymes and are resistant to separation from the cell wall (Claessen et al., 2003; Duong et al., 2012). There appears to be some redundancy among the chaplin proteins as only the simultaneous deletion of multiple chp genes attenuated aerial hyphae formation and reduced attachment to substrate surfaces (Claessen et al., 2004; de Jong et al., 2009). Like chaplins, rodlins are structural components of aerial mycelium in S. coelicolor (Claessen et al., 2002). Rodlin proteins are detected only in cultures forming aerial hyphae, and although disruption of rdlA and rdlB does not affect the formation of aerial structure, it does affect the formation of the rodlet layer (Claessen et al., 2002, 2004). Studies indicate that rodlins and chaplins interact to form the rodlet layer and that the rodlins weave chaplins into paired rodlets displaying a characteristic basketwork-like appearance on the surface of S. coelicolor spores (Wildermuth, 1970; Wildermuth et al., 1971; Claessen et al., 2002, 2004).
For differentiation into aerial hyphae, the substrate mycelium of S. coelicolor also requires SapB, a small, hydrophobic, morphogenetic peptide, to reduce surface tension at the colony interface on rich solid media such as R2YE (Willey et al., 1991, 1993, 2006; Tillotson et al., 1998; Wosten and Willey, 2000). Production of SapB requires the ram (rapid aerial mycelium formation) gene cluster ramCSAB (Willey et al., 1991; Ma and Kendall, 1994; Kodani et al., 2004). The regulatory gene ramR, immediately adjacent to ramB, plays a pivotal role in the regulation of SapB production, as RamR activates the ramC operon by binding directly to sequences upstream (Nguyen et al., 2002; O’Connor et al., 2002). Evidence suggests that SapB production is also influenced by the membrane localization of the precursor peptide RamS (Gaskell et al., 2012). It was proposed that both SapB and the chaplins are essential for normal aerial formation on rich media such as R2YE, whereas chaplins alone drive aerial morphogenesis on complex medium such as solid MS medium (Claessen et al., 2004; Capstick et al., 2007).
The regulation of development in streptomycetes is only partially understood, although bld and whi genes are known to be involved (Flardh and Buttner, 2009; Chater, 2011; McCormick and Flardh, 2011). bld mutations block the formation of aerial mycelium, resulting in a ‘bald’ appearance, and whi mutations block steps in the conversion of aerial mycelia to mature, gray spores, and hence whi mutants appear ‘white’ (Flardh and Buttner, 2009; Chater, 2011; McCormick and Flardh, 2011). A developmental regulatory cascade involving most bld and whi genes was established after years of study, with BldD apparently at the top of the regulatory cascade (Den Hengst et al., 2010; Bush et al., 2016). BldD is an auto-regulatory, DNA-binding protein (Elliot et al., 1998; Elliot and Leskiw, 1999), and its targets includes bldA, bldC, bldH, bldM, bldN, whiB, and whiG (Elliot et al., 2001; Den Hengst et al., 2010; Bush et al., 2016). To regulate its target genes, BldD needs to form a homodimer, a process that depends on c-di-GMP (Tschowri et al., 2014), and therefore BldD activity is essentially influenced by the level of this signal molecule (Tschowri et al., 2014). Recent research also provides evidence that collaboration between bld and whi factors (BldM and WhiI) or among whi factors (WhiA and WhiB) is required to control the key stages in Streptomyces development (Bush et al., 2013, 2016; Al-Bassam et al., 2014).
The genome of S. coelicolor encodes 67 paired two-component signal transduction systems (TCSs), 13 orphan response regulators, and 17 orphan sensor kinases (Bentley et al., 2002; Hutchings et al., 2004). Some TCSs have been characterized as regulators of primary or secondary metabolism (Hutchings et al., 2004); for example, PhoPR, the most studied TCS in S. coelicolor, is implicated in both phosphate metabolism and antibiotic production (Fink et al., 2002; Sola-Landa et al., 2003, 2005), whereas others are involved in secondary metabolism. Some TCSs have been implicated in development, such as BldM (Molle and Buttner, 2000), RamR (Keijser et al., 2002; Nguyen et al., 2002; O’Connor et al., 2002), and WhiI (Ainsa et al., 1999; Al-Bassam et al., 2014). However, the function of most of these signal systems and their roles in developmental regulation have not been fully explored.
SCO3012/3013 is one of the uncharacterized TCSs in S. coelicolor, and SCO3013 is annotated as the response regulator of this system. SCO3013 has 75% amino acid identify to MtrA (MtrAMTB) of Mycobacterium tuberculosis, which is an essential gene in that species (Zahrt and Deretic, 2000, 2001), and 69% amino acid identity to MtrA (MtrACGL) of Corynebacterium glutamicum, which is implicated in the regulation of cell wall metabolism and osmoprotection (Moker et al., 2004); therefore, SCO3013 was also named MtrA (Hoskisson and Hutchings, 2006). In S. coelicolor, mtrA (sco3013), its cognate sensor kinase gene mtrB (sco3012), and the flanking gene lpqB (sco3011), which encodes a putative lipoprotein, form an operon that is conserved in other actinobacteria, with MtrA displaying the highest sequence conservation of the three genes (Hoskisson and Hutchings, 2006). The critical biological role of MtrA in other gram-positive actinomycetes suggests that MtrA may have an important role in Streptomyces. Here, we report that MtrA is critical for development and that this role is conserved in streptomycetes.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The wild-type S. coelicolor and S. lividans strains and their derivatives were grown at 30°C on mannitol soya (MS) flour agar for spore production and conjugal transfer; on TSBY broth medium (3% tryptone soya broth, 10.3% sucrose, 0.5% yeast extract, final pH of 7.2) for mycelium preparation (Pospiech and Neumann, 1995); and on complex or rich medium (Kieser et al., 2000; Ou et al., 2009) media to observe morphological phenotypes. Various Escherichia coli strains were cultivated in Luria-Bertani (LB) liquid medium and used as indicated in Supplementary Table S1. Ampicillin (100 μg ml-1), apramycin (50 μg ml-1), thiostrepton (20 μg ml-1), kanamycin (25 μg ml-1), hygromycin (50 μg ml-1), chloramphenicol (25 μg ml-1), and nalidixic acid (25 μg ml-1) were added, as appropriate, to growth medium for selection of either E. coli or Streptomyces transformants.
Construction of the S. coelicolor M145 Genomic Library and Southern Blotting
Chromosomal DNA was isolated from S. coelicolor M145 mycelia grown in 50 mL TSBY broth containing 0.5% glycine, as described (Pospiech and Neumann, 1995). The genomic DNA was partially digested with MboI to yield DNA fragments of 35–45 kb, which were then ligated into the vector SuperCos I cosmid that had been previously linearized with XbaI, dephosphorylated, and digested with BamHI. The resulting ligation mixture was packaged into λ phage, followed by phage transfection into E. coli XL1-Blue MR using the MaxPlax Lambda Packaging Extracts Kit (Epicentre), according to the manufacturer’s instructions. Ampicillin-resistant colonies were grown in LB medium supplemented with ampicillin, and then stored in 25% glycerol at -80°C.
Primers 3013-hybrid-foward/reverse (Supplementary Table S2) that encompass the mtrA coding sequence were used to generate a 1065-bp DNA fragment, which was purified and labeled using reagents supplied in the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche); the labeled fragment was then used to probe a M145 cosmid library that had been transferred to nylon membranes using standard hybridization protocols (Sambrook and Russell, 2001). Finally, after addition of detection buffer, the membrane was exposed to X-ray film for 20 min to detect the chemiluminescent signal. Cosmid 19E7 generated a positive signal with the probe, and its insert was determined using inward-sequencing with primers specific to the cloning sites of the vector.
Deletion of mtrA from S. coelicolor M145
A PCR-targeting system was used to delete mtrA from S. coelicolor essentially as described (Gust et al., 2003). Briefly, a disruption cassette was generated by PCR using pIJ773 as template with primer pair mtrA-Target-F/R (Supplementary Table S2), and then electro-transformed into E. coli BW25113, which contains pIJ790 that provides the λ-Red system to enhance recombination, and cosmid 19E7, which contains the mtrA gene, to generate the mtrA-deleted cosmid ΔmtrA::aac(3)IV. Deletion of mtrA was verified by both PCR using primer pair ΔmtrA-Confirm-F/R (Supplementary Table S2) and by restriction enzyme digestion analysis. Cosmid ΔmtrA::aac(3)IV was then introduced into the non-methylating E. coli strain 12567/pUZ8002 before transferring into S. coelicolorM145 by conjugation. After several rounds of selection on MS agar containing apramycin, the deletion of mtrA from the apramycin-resistant exconjugant ΔmtrA was confirmed by PCR using the primer pair ΔmtrA-Confirm-F/R.
Owing to the high sequence identity (99–100%) between sli_3357 of S. lividans and mtrA of S. coelicolor and the strong sequence similarity between their flanking sequences, the same mutated cosmids were used to generate mutant Δsli_3357 in S. lividans 1326.
Deletion of sven_2756 from S. venezuelae
To mutate sven_2756 in S. venezuelae, a 2108-bp flanking sequence at the left side of the gene was used as the left arm for homologous recombination and was amplified using primer pair SVEN2756-L Forward/Reverse (Supplementary Table S2), which carry an XbaI and SpeI site, respectively. The 2108-bp DNA fragment was then cloned into pEasy-Blunt to generate p-LArm. A 2071-bp flanking sequence at the right side of the gene was used as the right arm for homologous recombination and was amplified using primer pair SVEN2756-R Forward/Reverse (Supplementary Table S2), which carry an SpeI site and a HindIII site, respectively; this fragment was also inserted into pEasy-Blunt to generate p-RArm. A third fragment, 1269-bp in length and containing a kanamycin cassette, was amplified using primer pair Kana-Forward/Reverse and cloned into pMD18-T. After sequence verification, the right arm was released from p-RArm by SpeI and HindIII digestion, ligated with p-LArm pretreated with HindIII and SpeI, and the resulting plasmid pL-R was verified by digestion with HindIII and XbaI. Next, the kanamycin cassette was removed by SpeI digestion, purified, and ligated with pL-R pretreated with SpeI (to separate the left and the right arm); the resulting plasmid pL-K-R was verified by digestion with HindIII and XbaI. Then, the fragment containing the left and right arms and the resistance cassette was released by HindIII and XbaI digestion, purified, and inserted into pJTU1278 pretreated with HindIII and XbaI. The resulting plasmid was verified by restriction analysis and was designated pMu-2756.
pMu-2756 was introduced into the non-methylating E. coli strain 12567/pUZ8002, and the transformants were used as donor strains in conjugation. The protocol for conjugation and selection of mutants was essentially as described for S. coelicolor M145, except that R2-S medium was used for conjugation as described (Bibb et al., 2012). After several rounds of selection on R2S agar containing kanamycin, the deletion of sven_2756 from the exconjugant was confirmed by PCR using the primer pair SVEN2756-C Forward/Reverse (Supplementary Table S2).
Genetic Complementation
To generate a complemented strain for ΔmtrA, the upstream intergenic region and coding sequence of mtrA or mtrA and mtrB were amplified by PCR, using corresponding primer pairs (Supplementary Table S2). The amplicon was purified, inserted into pCR-Blunt, and following sequencing verification, the insert was excised and ligated with the pre-cut integrating plasmid pMS82 (Gregory et al., 2003), which confers hygromycin resistance, to generate pCom-3013SCO and pCom-3012/13SCO, respectively. Also based onpMS82, pCom-2757SV EN was constructed, which contains the upstream intergenic sequence and the coding sequence of sven_2756 of S. venezuelae. All plasmid constructs were verified by sequencing and were introduced into mutant strains by conjugation to generate the complemented strains (Supplementary Table S1). Exconjugants with resistance to hygromycin were screened and confirmed by PCR analysis.
RNA Isolation, Reverse Transcription-PCR (RT-PCR), and Real-time PCR
To extract RNA, wild-type and mutant strains of Streptomyces were grown at 30°C on YBP solid medium covered with plastic cellophane, and the mycelia were collected at various times, ground in liquid nitrogen, and then dispensed into Rezol reagent (SBSBIO). Crude RNA samples were treated twice with ‘Turbo DNA-free’ DNase reagents (Ambion) to remove chromosomal DNA, and reverse transcription was carried out, as described (Zhang et al., 2014). The SYBR Premix Ex Taq (TaKaRa) was used under recommended conditions on a Roche LightCycler480 thermal cycler to determine the melting curve of PCR products and their specificity, and for real-time PCR assays. Relative quantities of cDNA were normalized for the hrdB gene, which encodes the major sigma factor of Streptomyces. Sequences of primers used in RNA analysis are listed in Supplementary Table S2.
RNA-Seq and Data Analysis
For RNA-Seq analysis, the total RNA of S. coelicolor M145 and ΔmtrA was extracted from cultures incubated at 30°C for 72 h on solid YBP medium covered with cellophane and were then treated with RNase-free DNase I (Invitrogen) twice according to the recommended protocols. Two sets of RNA for both strains were prepared separately. The integrity of total RNA was determined using a Thermo NanoDrop, and the RNA Integrity Number value of each sample met the standard required for preparing a cDNA library. The cDNA libraries were prepared according to the manufacturer’s instructions (Illumina). Briefly, rRNA in 1 μg total RNA was depleted using Ribo-zero rRNA Removal solution (Illumina), leaving only mRNA, and the mRNA was then fragmented into small pieces in fragment mix at elevated temperature to avoid priming bias when synthesizing cDNA. Using random primers and Super Script II (Invitrogen), the small mRNA fragments were then converted into double-stranded cDNA by first- and second-strand cDNA synthesis, followed by end-repair and 3′-adenylation to produce cDNA fragments with a single ‘A’ base overhang at their 3′-ends; adapters were then ligated to the ends of the cDNA fragments. Fifteen rounds of PCR amplification were performed to enrich the adapter-modified cDNA library using primers complementary to the ends of the adapters, and PCR products were purified using Ampure XP beads (Agencourt). The concentration of the cDNA library was determined with a Taqman probe using ABI StepOnePlus Real-time PCR system, and the size range of the library insert was determined with Agilent DNA 1000 Reagents using the Agilent 2100 Bioanalyzer. The cDNA library products were denatured into single strands with the addition of NaOH, diluted to optimal concentration, and loaded into a FlowCell to hybridize with the adapter fixed on the FlowCell. Bridge-PCR amplification was performed with TruSeq PE Cluster Kit V3-cBot-HS (Illumina), and the FlowCell samples were sequenced using HiSeq2000 (Illumina).
The expression level of each gene was normalized by the number of reads per kilobase of transcriptome per million mapped reads (RPKM). The cut-off value for determining gene transcriptional activity was determined based on a 95% confidence interval for all RPKM values of each gene.
Construction of an mtrA Expression Plasmid and Purification of Protein MtrA
The mtrA coding sequence was amplified using primers mtrA-Exp-F (with an NdeI adaptor) and mtrA-Exp-R (with a HindIII adaptor) (Supplementary Table S2), and the PCR product was purified by agarose gel electrophoresis and inserted into pMD18-T (Takara). After sequence verification, the inserts were excised by NdeI and HindIII digestion, gel-purified, and ligated into NdeI/HindIII-cut pET28a (Invitrogen) to generate pEX-mtrA, which was used to transform E. coli Rosetta(DE3)pLysS (Novagen). Expression of MtrA was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (1.0 mM) when the cell density was around 0.6 (at OD600 nm), with incubation for 4–5 h at 30°C. Cell lysates were prepared by sonication in binding buffer (50 mM NaH2PO4, 250 mM NaCl, 20 mM imidazole, pH 8.0), and the His-tagged MtrA was purified using Ni-NTA-Sefinose Column (Sangon.), using washing buffer (50 mM NaH2PO4, 250 mM NaCl, 40 mM imidazole, pH 8.0) and then elution buffer (50 mM NaH2PO4, 250 mM NaCl, 250 mM imidazole, pH 8.0). Purified protein was then dialyzed in dialysis cassettes (10,000 MWCO, Thermo Scientific) in a dialyzing buffer (50 mM NaH2PO4, 50 mM NaCl, pH 8.0) before concentrating with centrifugal filters (10,000 MWCO, Millipore). Protein concentration was determined using the bicinchoninic acid assay (Pierce).
Scanning Electronic Microscopy (SEM)
Briefly, spores of S. coelicolor M145 and its derivative strains were inoculated onto MS agar medium; for S. venezuelae strains, YBP agar medium was used instead. A sterile coverslip was inserted into the agar at an angle to allow the culture to overgrow its surface. After 5 days of incubation at 30°C, the coverslip was removed, fixed with fresh 2% glutaraldehyde (pH 7.2) for 2 h at 30°C, and washed three times with 0.1 M PBS buffer (pH 7.0) before treating with 1% osmic acid. The coverslips were then dehydrated by soaking in a series of ethanol gradients, dried in a Leica EM CPD300 Critical Point Dryer, coated with gold in a Cressinton Sputter Coater 108, and examined with a scanning electron microscope (FEI Quanta250 FEG, United States).
Electrophoretic Mobility Shift Assays (EMSAs)
DNA probes containing the upstream region of selected genes were generated by PCR amplification using specific primer pairs as listed (Supplementary Table S2). For short probes containing putative MtrA binding sequence, complementary oligonucleotides were annealed. The DNA probes were labeled with biotin-11-UTP using the Biotin 3′ End DNA Labeling Kit (Thermo Scientific), according to the manufacturer’s instruction, and 50 fmol of labeled probes were mixed with differing amounts of purified MtrA in binding buffer (20 mM Tris-HCI, 2 mM EDTA, 20 mM KCI, 0.5 mM DTT, 4% Ficoll-400, pH 8.0). For an unspecific competitor, 2 μg of poly (dI-dC) was included in the reactions (20 μl total reaction volume). The DNA and protein mixes were incubated at room temperature for 15 min, and then were separated on 8% non-denaturing polyacrylamide gels. After gel separation, the DNA was transferred to and fixed on nylon membrane, and then blocked, washed, and processed before signal detection using the ECL Western Blotting Analysis System kit (GE Healthcare) and exposure to X-film.
DNaseI Foot-Printing Assay
The DNA probes used in electrophoretic mobility shift assays (EMSA) were cloned into pMD18-T to generate plasmid templates. For preparation of fluorescent FAM-labeled probes, the promoter region of each gene was PCR amplified with Dpx DNA polymerase (TOLO Biotech) from the plasmid template using primers M13F-47 (FAM) and M13R-48. The FAM-labeled probes were purified by the Wizard® SV Gel and PCR Clean-Up System (Promega) and were quantified with NanoDrop 2000C (Thermo). DNase I footprinting assays were performed essentially as described (Wang et al., 2012). For each assay, 400 ng of probe was incubated with different amounts of purified MtrA in a total volume of 40 μl, which also included poly (dI-dC) as the unspecific competitor. After incubation for 30 min at 25°C, 10 μl of solution containing approximately 0.015 units DNase I and 100 nmol freshly prepared CaCl2 was added, followed by further incubation for 1 min at 25°C. The reaction was stopped by adding 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). Samples were extracted with phenol/chloroform, precipitated with ethanol, and the pellets were dissolved in 30 μl MiniQ water. Preparation of the DNA ladder, electrophoresis, and data analysis were performed as described previously (Wang et al., 2012; Cao et al., 2015), except that the GeneScan-LIZ500 size standard (Applied Biosystems) was used.
Identification of an MtrA Consensus Recognition Sequence
The sequences protected or shifted by MtrA in the regions upstream of sco1189, sco1489, sco1568, sco2136, sco2210, sco3485, sco3561, sco3863, sco5583, sco6029, sco7434, and sco7458 were used as input for the MEME software tool (Bailey and Elkan, 1994) for the MtrASCO motif search.
Results
Disruption of mtrA Leads to a Typical Bald Phenotype in S. coelicolor
We screened a transposon library of S. coelicolor M145 for mutants with altered production of pigmented antibiotics (Xu et al., 2017), discovering four mutants with the bald phenotype typical of strains lacking aerial hyphae; these mutants contained transposons within the coding sequence of mtrA, suggesting a role for MtrA in developmental control. To verify the effects of MtrA on Streptomyces morphology, we generated the deletion mutant ΔmtrA, which lacks the entire mtrA coding sequence, in strain S. coelicolor M145. ΔmtrA exhibits significant morphological differences from strain M145 when grown on various media (Figure 1A and data not shown). On MS medium, formation of aerial mycelia was severely delayed in ΔmtrA, with none observed even after 8 days of growth (Figure 1A), in sharp contrast to M145, which began formation of aerial mycelia as early as 48 h under the test conditions. However, after prolonged incubation on MS medium (∼10 days), ΔmtrA was capable of developing aerial mycelia, with mature spores forming after approximately 2 weeks. Notably, on R2, R5, R2YE (Kieser et al., 2000) and YBP (Ou et al., 2009), ΔmtrA maintained a ‘permanent’ bald phenotype, with no aerial mycelia observed even after weeks of growth, whereas wild-type M145 began forming aerial mycelia at 48 h under the same conditions. Overall, ΔmtrA displayed a striking morphological change, i.e., a bald phenotype that was conditional upon the growth medium, a very similar phenotype to that reported for SapB- and chaplin-deficient mutants and bld mutants (Capstick et al., 2007; Chater, 2011). We further observed that ΔmtrA cultures appeared very ‘wet’ on rich media and that water droplets spread easily over the ΔmtrA culture surface, suggesting that, compared to the wild-type strain, this mutant has a much more hydrophilic nature.
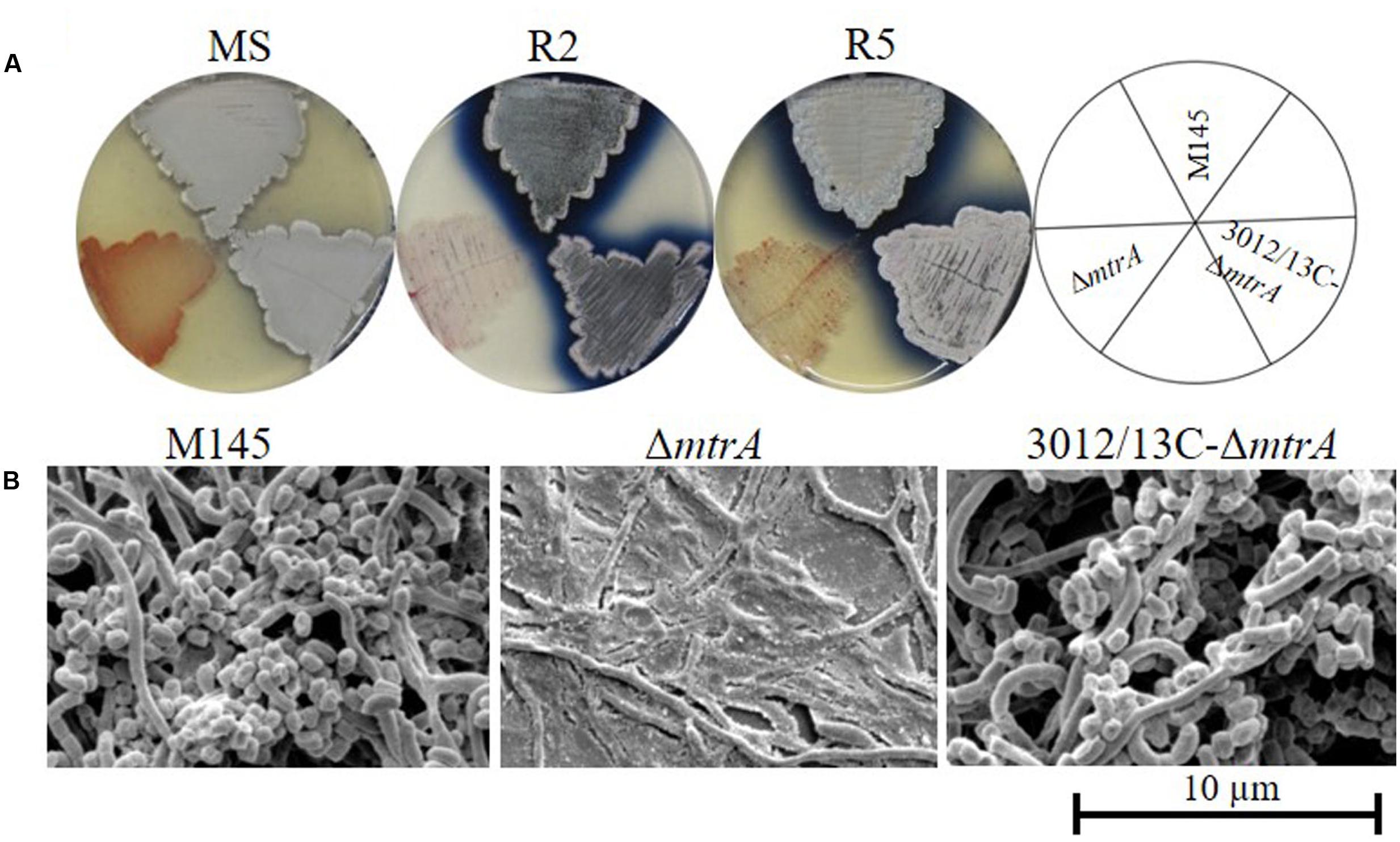
FIGURE 1. Requirement of mtrA for the formation of aerial hyphae in S. coelicolor. (A) Phenotypes of the S. coelicolor wild-type strain M145, the mtrA deletion mutant ΔmtrA, and the mtrA-complemented strain C-ΔmtrA grown at 30°C on solid MS (7 days), R2 (6 days), and R5 (6 days) media. (B) SEM images of M145, ΔmtrA, and C-ΔmtrA after growth on MS agar for 5 days. Images reveal the classic ‘bald’ phenotype for the deletion mutant and normal aerial hyphae for the wild-type and complemented strains. The scale bar is 10 μm.
To confirm that the profound phenotypic changes in ΔmtrA were directly due to mutation of mtrA, we introduced plasmid pCom-3012/13SCO, which contains the coding sequence of sco3012 (mtrB) and sco3013 (mtrA) and the upstream intergenic sequence, into ΔmtrA to generate the complemented strain 3012/13C-ΔmtrA; this complementation restored the growth of aerial mycelium and spore formation to a level comparable to that of wild-type M145 (Figure 1A), confirming a critical role for mtrA in developmental control in S. coelicolor. However, when pCom-3013SCO, which contains the coding sequence of sco3013 (mtrA) and the upstream intergenic sequence, was introduced into ΔmtrA, the phenotype was only partially complemented in strain 3013C-ΔmtrA under most conditions tested. As there is only one nucleotide space between the putative start codon of mtrB and the annotated stop codon of mtrA, and as the entire coding sequence of mtrA, except for the start and stop codons, was replaced by an apramycin cassette, translation of MtrB may be impaired in ΔmtrA. However, the transcription of mtrB did not appear to be affected by the mtrA mutation, as inferred from RNA-Seq data in which mtrA was the most down-regulated gene due to the absence of the coding sequence, whereas no obvious change was detected for mtrB. It is possible, though, that the inefficient complementation of ΔmtrA by mtrA alone (pCom-3013SCO) compared to mtrAB (pCom-3012/13SCO) under some conditions, such as growth on R2 and R5 media, might be due to a polar effect of the mtrA mutation. However, the fact that under other conditions, such as growth on MS medium, mtrA complements the deletion nearly as well as mtrAB argues that any such polar effect is not strong. Nevertheless, the results from this and other studies indicate that MprB is required for maintaining the full activity of MprA (Som et al., 2017), consistent with MprA and MprB composing a two-component system.
Morphological Analysis by Scanning Electron Microscopy (SEM)
In other Streptomyces mutants, the bald phenotype is associated with the absence of aerial mycelia. Therefore, the morphology of ΔmtrA was visualized by scanning electron microscopy (SEM) (Figure 1B), using strains grown for 5 days on solid MS medium. Both the wild-type M145 and the complemented strain 3012/13C-ΔmtrA were abundantly covered with loosely coiled spore chains, whereas ΔmtrA was covered only by clumps of submerged mycelia (Figure 1B), confirming that ΔmtrA is defective in the formation of the aerial mycelium, and indicating an essential role for MtrA in normal morphological development.
Differential Expression of Developmental Genes in ΔmtrA
To investigate the mechanisms underlying the bald phenotype of ΔmtrA, we conducted RNA-Seq analysis on wild-type M145 and ΔmtrA, using cultures grown for 72 h on solid YBP medium. All eight chaplin genes were significantly down-regulated in ΔmtrA compared to M145, as were the two rodlin genes, rdlA and rdlB (Figure 2A). ramCSAB and ramR, involved in SapB production, were also down-regulated in the mutant (Figure 2B). Interestingly, bldD, a global transcriptional regulator in S. coelicolor (Elliot et al., 1998, 2001; Den Hengst et al., 2010), was overexpressed in ΔmtrA (Figure 2C), whereas the bldK operon, which is responsible for the export of a signal molecule critical for development (Nodwell et al., 1996), was significantly down-regulated, suggesting that MtrA represses bldD but activates bldK. In addition, whiH and whiI, the two whi genes that play an important role in the late stages of differentiation (Ryding et al., 1998; Ainsa et al., 1999), were down-regulated in ΔmtrA (Figure 2C), suggesting that MtrA activates whiH and whiI under the conditions tested. Overall, RNA-Seq analysis indicated that MtrA not only affects genes required for the formation of aerial mycelium but also genes dedicated to spore maturation.
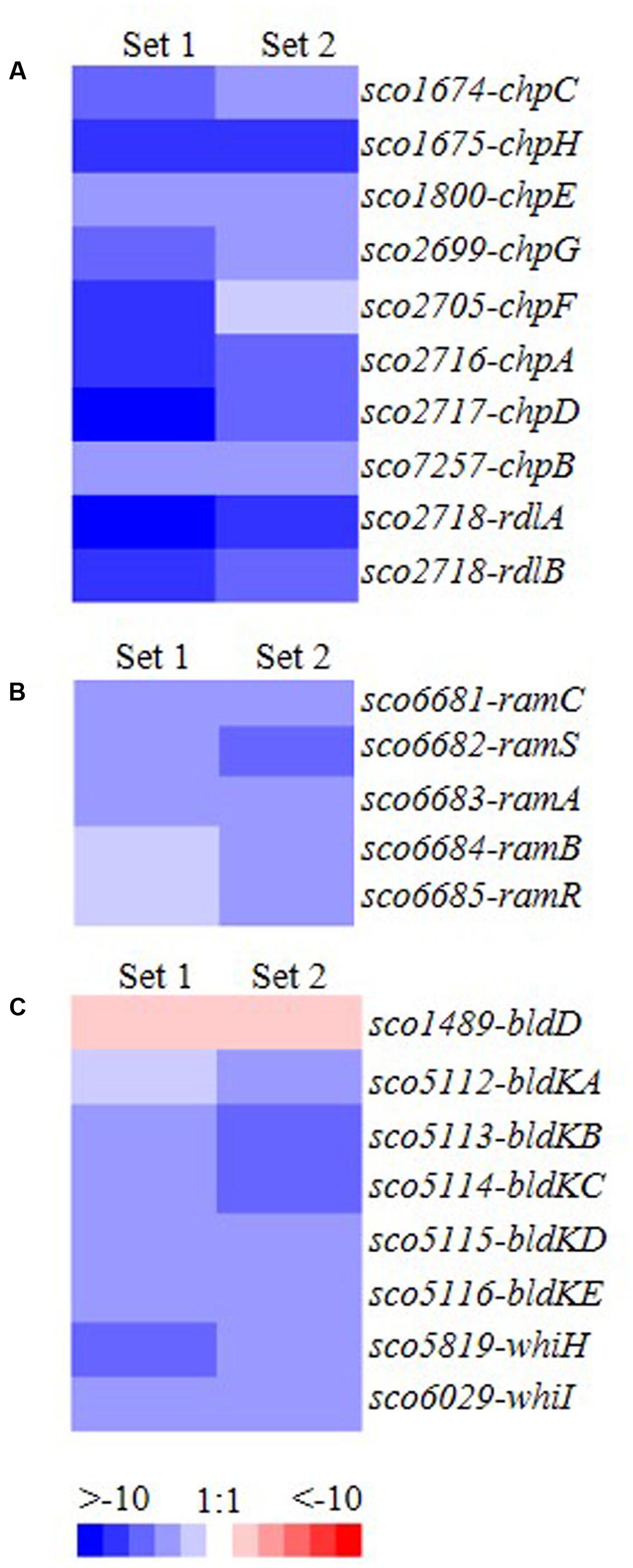
FIGURE 2. Transcriptomic analysis of genes involved in cellular development and whose expression differs between ΔmtrA and wild-type M145. (A) chp and rdl genes (involved in formation of the hydrophobic sheath of the aerial mycelium); (B) ram genes (involved in production of SapB); (C) developmental regulatory genes of the bld and whi families. Total RNA was extracted from mycelia of M145 and ΔmtrA grown for 72 h at 30°C on YBP solid agar, cDNA libraries were prepared, and transcript levels were determined by RNA-Seq. The value for a specific gene was calculated as the log2 ratio of the expression level in RPKM (reads per kilobase transcriptome per million mapped reads) of the gene in ΔmtrA relative to that in M145, with red indicating an increase in transcript abundance in ΔmtrA and blue indicating a decrease.
Analysis of the Expression Patterns of chp and rdl Genes in ΔmtrA during Development
The RNA-Seq analysis was conducted at 72 h of growth and was thus a snapshot of the gene expression patterns occurring during the relatively long developmental process. For a more comprehensive view of the role of MtrA during different developmental phases and to validate the RNA-Seq data, real-time PCR was performed. RNA was extracted from M145 and ΔmtrA at 12-h intervals over 36–84 h of growth on solid YBP medium, a time period that covers the formation of vegetative mycelium, aerial mycelium, and spores. The expression level of each gene at 36 h in M145 was arbitrarily set to 1. Based on their expression patterns, the eight chaplin genes could be divided into two groups (Figure 3A). The first group, comprising chpB (sco7257), chpC (sco1674), chpE (sco1800), chpF (sco2705), chpG (sco2699), and chpH (sco1675), demonstrated peak levels of transcription in M145 between 60 and 72 h, with maximal fold changes in M145 ranging from 3.94 ± 0.44 for chpE to 18.96 ± 4.71 for chpB, with transcript levels decreasing thereafter. chpC, chpH, and chpE showed the most similar patterns, peaking at 60 h in M145, albeit the induction levels of chpE were less than half that of the other two genes (Figure 3A). In ΔmtrA, expression of genes in this first group was markedly lower, with transcription of chpC and chpH barely detectable at each of the five time points (Figure 3A), indicating that MtrA strongly influences these genes throughout development. The second group of chp genes, chpA (SCO2716) and chpD (SCO2717), displayed dramatically increasing expression levels throughout the development of M145, with peak expression levels at the last time point (84 h) that were approximately 148.2 and 165.5 times greater than those at 36 h for these genes (Figure 3A). However, similar to the genes in group 1, the induction of chpA and chpD was abolished in ΔmtrA (Figure 3A).
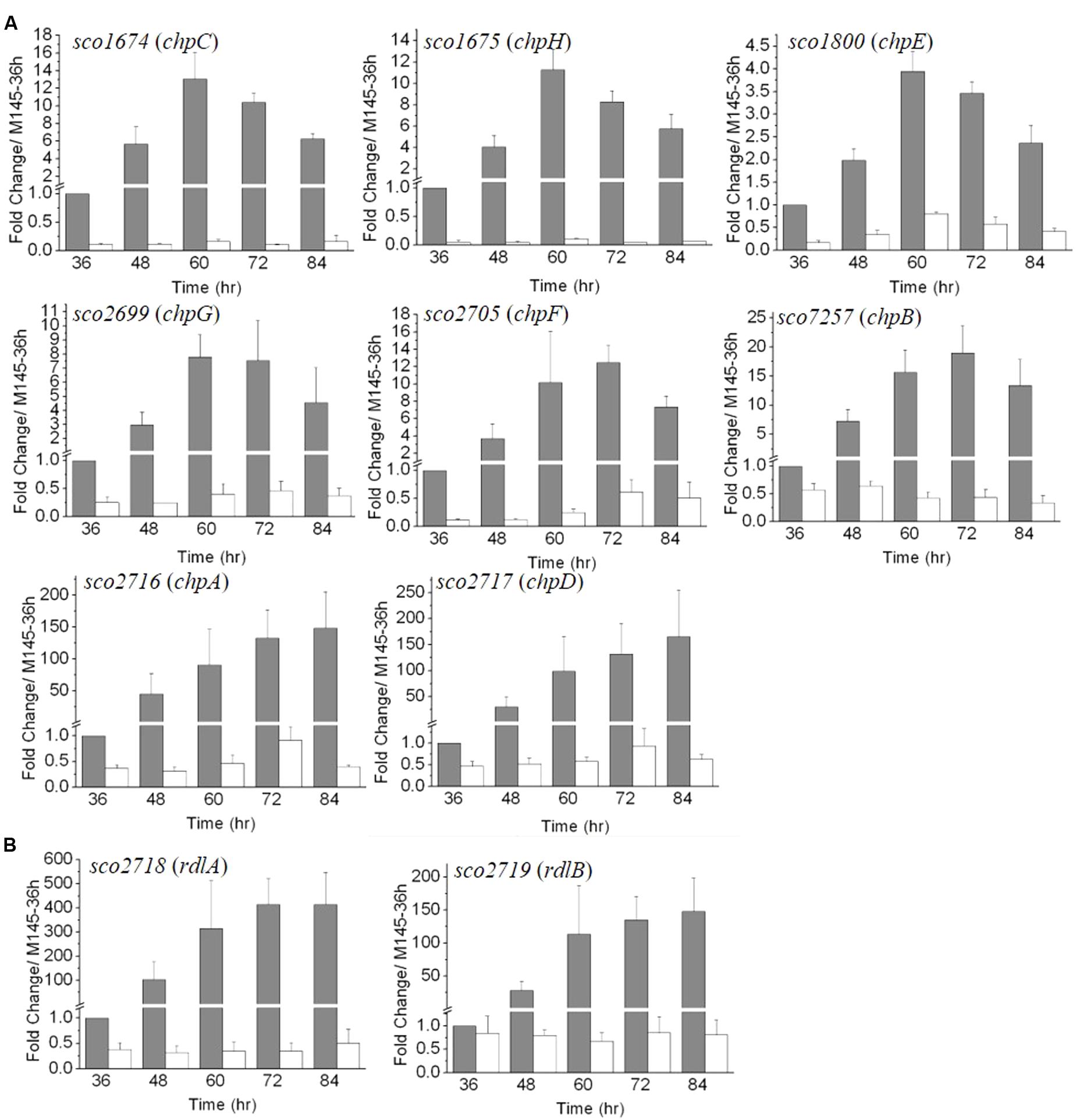
FIGURE 3. The developmentally regulated expression of chaplin (A) and rodlin (B) genes in S. coelicolor M145 is blocked in ΔmtrA. Expression of these morphogen genes was determined by real-time PCR analysis. M145 and ΔmtrA were grown on YBP solid medium, and RNA samples were isolated at the indicated times. Expression of hrdB, encoding the major sigma factor in M145, was used as an internal control. For each gene, the expression level in M145 at 36 h was arbitrarily set to one. The y-axis shows the fold change in expression levels in M145 (gray bars) and ΔmtrA (light bars) over the levels in M145 at 36 h. Results are the means (± SD) of triplet experiments.
In M145, rdlA and rdlB displayed transcription patterns similar to those of chpA and chpD (Figure 3B), with highest transcription levels detected at the last time points (Figure 3B). Although rdlA and rdlB are divergently transcribed, rdlA was induced more highly than rdlB at each time point, culminating in fold changes of 414.1 ± 105.8 and 147 ± 51.3 at the last time point for rdlA and rdlB, respectively. However, induction of both genes was blocked in ΔmtrA, with expression values of less than 1.0 at all time points (Figure 3B), indicating a dramatic effect of MtrA on the developmentally regulated expression of rdl genes.
Transcription of ram Genes in ΔmtrA during Development
SapB is essential for aerial hyphae formation on rich medium (Willey et al., 1991, 1993), and so, to investigate the impact of MtrA on the expression of genes involved in SapB production, transcription of the ram cluster was examined using the same set of RNA samples described above. Whereas the chp and rdl genes were more highly induced during the late developmental phases when aerial mycelia and spores start to form, ramC expression was highest at the first time point (36 h), with expression decreasing to ≤0.2 of the control level for the remaining time points in M145 (Supplementary Figure S1); this finding is in agreement with a previous report, which demonstrated that RamC peaks at 36 h in the wild-type strain when grown on R2YE medium (O’Connor et al., 2002). However, in ΔmtrA, transcription of ramC was barely detectable at any of the five time points (Supplementary Figure S1). The temporal expression profiles of ramS, ramA, and ramB were similar to those of ramC in M145 and ΔmtrA (Supplementary Figure S1), with the consistently decreased expression in the mutant providing evidence that these genes form an operon in S. coelicolor and that MtrA is essential for their activation, especially at the very early phase of development.
As the ramC operon is under the direct control of RamR (Nguyen et al., 2002; O’Connor et al., 2002), we also compared transcription of ramR in M145 and ΔmtrA. ramR expression showed only minor changes in M145, and its expression was only slightly lower in ΔmtrA. For example, in ΔmtrA at 36 h, ramR expression levels were about half the levels found in M145 (Supplementary Figure S1), whereas at the same time point, ramC was down-regulated nearly 50-fold in ΔmtrA (Supplementary Figure S1). These findings suggest that the dramatically decreased expression of ramCSAB is more likely due to the absence of MtrA rather than to the slight reductions in ramR expression, and therefore that, compared to RamR, MtrA has a more dominant regulatory effect on ramCSAB under the conditions tested.
Transcription of bld Genes in ΔmtrA during Development
Our above data indicated that expression of genes involved in the formation of aerial mycelium was blocked in ΔmtrA. Notably, these genes are also poorly expressed in most bld mutants (Willey et al., 1991; Elliot et al., 2003; Bibb et al., 2012), and our RNA-Seq analysis had indicated that bldD and bldK genes were differentially expressed in ΔmtrA at 72 h growth (Figure 2). To determine if the expression of additional bld genes was altered in ΔmtrA, we used real-time PCR. For the majority of bld genes, transcription was either not altered over the five time points examined (bldA, bldB, and bldM) or only slightly reduced at certain time points (bldC, bldG, bldH, and bldN) by deletion of mtrA (Supplementary Figure S2), suggesting a generally minor effect of mtrA on their expression. In contrast, bldD and genes of the bldK operon exhibited markedly different expression between M145 and ΔmtrA throughout development (Figures 4A,B). The bldK operon encodes an oligopeptide permease, which transports a signal molecule that begins the signaling regulatory cascade required for aerial mycelium formation (Nodwell et al., 1996). bldKA, the first gene of the operon, showed a gradually decreasing expression pattern (Figure 4A), very similar to that of ramC in M145 (Supplementary Figure S1), from the peak level at 36 h to the lowest level of 0.33 ± 0.031 at 84 h, in agreement with the role of BldK during the early developmental stage. However, consistent with the RNA-Seq analysis, no induction of bldKA was detected in ΔmtrA at any time point, resulting in a 34.5-fold (1 versus 0.029 ± 0.007) difference at 36 h between M145 and the ΔmtrA mutant (Figure 4A). The expression patterns of bldKB and bldKC in the two strains were similar to that of bldKA (Figure 4A). Our data indicate that MtrA activates bldK, especially at the early phase of development.
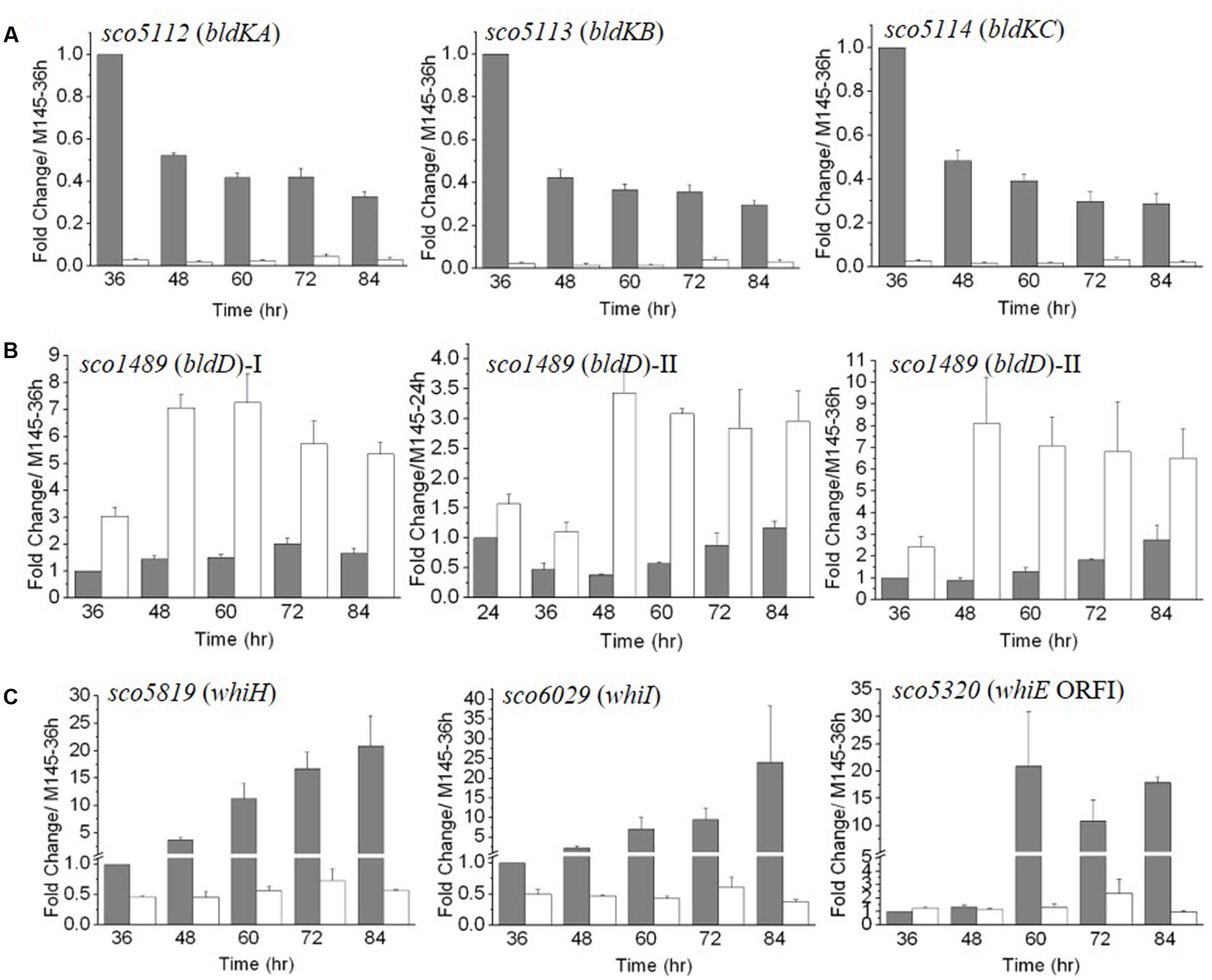
FIGURE 4. The temporal expression of bld (A,B) and whi (C) genes in ΔmtrA. Gene expression in M145 and ΔmtrA was determined by real-time PCR analysis. RNA samples were isolated from M145 and ΔmtrA and analyzed as described for Figure 3. The y-axis shows the fold change in expression level in M145 (gray bars) and ΔmtrA (light bars) over the level of M145 at 36 or 24 h (arbitrarily set to one). Results are the means (± SD) of triplet experiments.
In M145, transcription of bldD was fairly constant, ranging from 1.0 to 2.02 ± 0.21 times the level at 36 h (Figure 4B); however, its expression was notably higher in ΔmtrA at each of the five time points, from 36 to 84 h, with a maximal fold change of 7.26 ± 1.08 for ΔmtrA at 60 h versus only 1.50 ± 0.12 for M145 at the same time, suggesting that MtrA represses bldD at these time points. Expression of bldD normally peaks at the early growth stage (Elliot et al., 1998, 2001; Den Hengst et al., 2010); thus, a second set of RNA samples that included an earlier time point (24 h) was prepared, and expression of bldD in the two strains was evaluated relative to the bldD expression level at 24 h in M145 (Figure 4B, middle panel). In M145, expression of bldD at 24 h was approximately twofold higher than at 36 h, consistent with previous reports (Elliot et al., 1998; Den Hengst et al., 2010). In ΔmtrA, expression of bldD was consistently higher than in M145 at each of the six time points tested and maintained similar, high levels from 48 to 84 h (Figure 4B). When the bldD expression level in M145 at 36 h was used as the control for the second set of RNA samples, a similar expression pattern was observed as for the first set of RNA samples (Figure 4B, compare left and right panels for bldD), confirming that MtrA represses bldD, especially at the late developmental phase, and revealing MtrA as the first identified regulator of bldD other than BldD itself.
Transcription of whi Genes in ΔmtrA during Development
Our preliminary transcriptomic data indicated that two whi genes, whiH and whiI, were downregulated in the late stages of development in ΔmtrA (Figure 2C). For a more comprehensive view of the role of MtrA in whi expression, we examined the transcription of the eight canonical whi genes throughout development. Transcription of whiA, whiB, whiG, and whiJ was not altered, and whiD was slightly upregulated in ΔmtrA at the five time points tested (Supplementary Figure S3). In contrast, whiH and whiI showed developmentally associated induction in M145, with peak values at the last time point of 84 h (Figure 4C), consistent with a previous report (Ryding et al., 1998; Ainsa et al., 1999). In S. coelicolor, whiH and whiI are critical for cell differentiation (Ryding et al., 1998; Ainsa et al., 1999), and notably, their induction was absent in ΔmtrA (Figure 4C), suggesting that MtrA has a positive role in the developmentally regulated expression of whiH and whiI.
The whiE gene cluster, responsible for the synthesis of the gray pigment of mature spores, contains eight ORFs, of which whiE-ORFI through whiE-ORFVII appear to compose an operon that is divergently transcribed from whiE-ORFVIII (Davis and Chater, 1990; Kelemen et al., 1998). We chose whiEORF1 and ORFVIII as representatives of the cluster, and in M145, found that these ORFs showed a similar, marked increase in expression at 60 h, with levels remaining well-above control levels through the later time points (Figure 4C and Supplementary Figure S3). This induction was not detected in ΔmtrA, suggesting that MtrA is required for whiE expression, consistent with our observation that the spores of ΔmtrA lacked the gray color typical of M145 spores.
In summary, based on our extensive real-time PCR analyses, MtrA not only affects expression of the structural components of the aerial mycelium but also two classes of regulatory genes known to control developmental processes in S. coelicolor.
Binding of MtrA to the Upstream Regions of Developmental Regulatory Genes
Our above analysis indicated that MtrA controls multiple developmental regulatory genes. To investigate whether MtrA directly regulates these genes, the sequences upstream of bldD, bldKA, whiE-ORF1, and whiI were amplified using specific primer pairs (Supplementary Table S2). The resulting DNA fragments, ranging from 250 to 300 bp, were purified, labeled with biotin-11-UTP, and used as EMSA probes. Purified MtrA shifted DNA probes containing the sequences upstream of bldD and whiI but not those upstream of bldKA or whiE-ORF1 (Figure 5A). These in vitro analyses suggested that MtrA interacts directly with the promoters of two critical developmental regulatory genes.
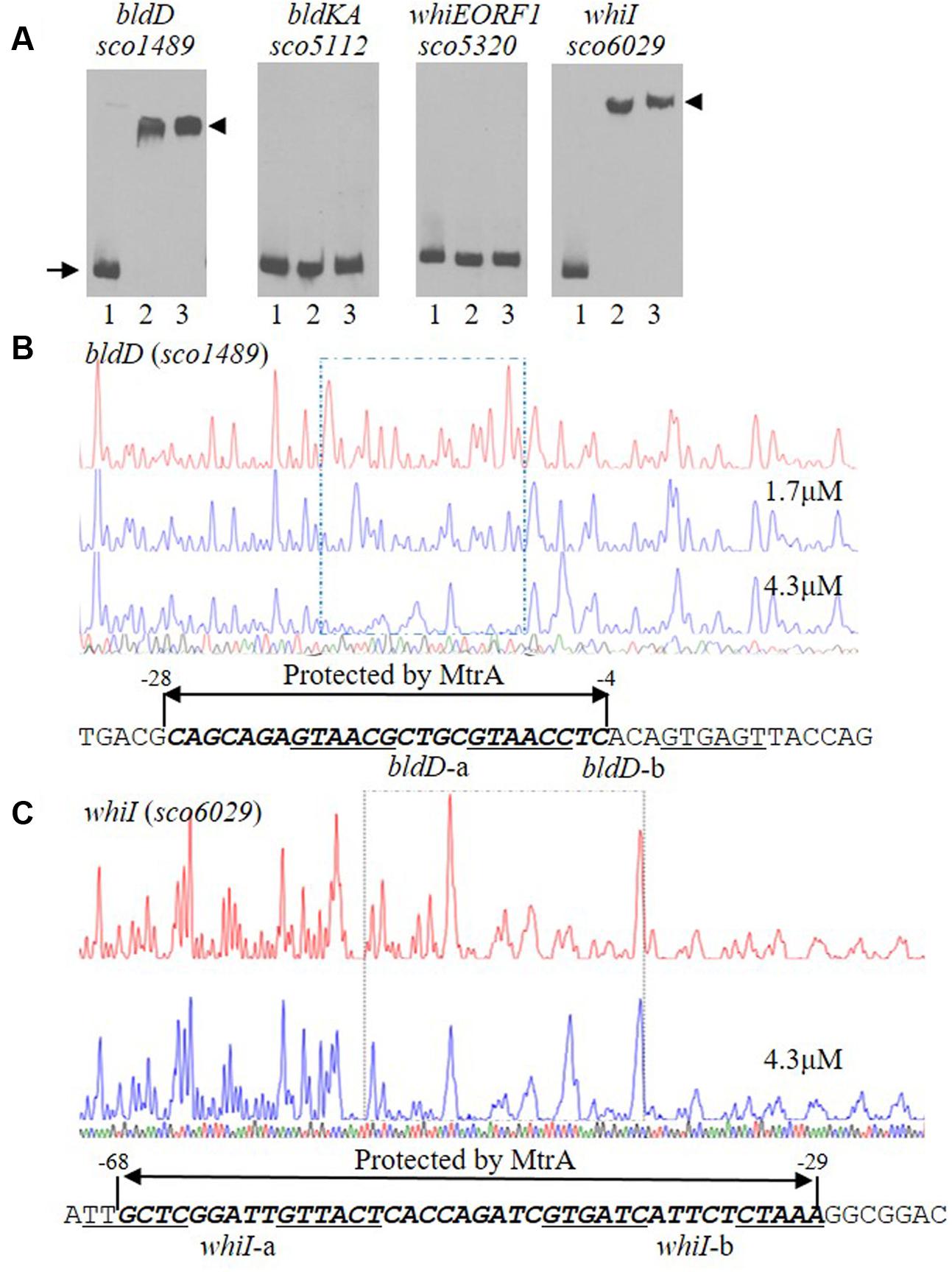
FIGURE 5. Interactions of MtrA with the upstream regions of bldD and whiI. (A) For EMSAs, a fixed amount of labeled probe containing the intergenic region upstream of the indicated gene was incubated in reactions containing no MtrA (lane 1), or 1.6 or 3.2 μg MtrA (lanes 2 and 3, respectively). The DNA fragments used as probes were amplified by PCR. The length of the probe for each upstream region is: bldD (282 bp), bldKA (265 bp), whiE-ORF1 (267 bp), and whiI (293 bp). The arrow indicates the position of free probes, and arrowheads indicate the position of the shifted probes. DNase I footprinting assays of the coding strand of bldD (B) and whiI (C). The upper electropherograms in red are the control reactions without addition of MtrA protein. The reactions including MtrA are shown below in blue. The MtrA-protected regions are indicated by the dotted rectangles. The superimposed electropherograms shows the sequencing reactions. The nucleotide sequence of the MtrA-protected region for each gene is marked by bold italics and a double arrow, and the numbers indicate position relative to the TSP of each gene. The potential MtrA sequence is underlined.
Detection and Validation of the MtrA Binding Sequence
To map the precise sequences protected by MtrA, we performed DNase I footprinting assays using FAM-labeled probes. For bldD, the region from -276 to +6 with respect to its TSP (Elliot et al., 1998; Elliot and Leskiw, 1999) was used as template, and MtrA protected the sequence from -28 to -4 in the sense strand (Figure 5B), which overlaps the -10 region (Elliot et al., 1998; Elliot and Leskiw, 1999). For whiI, we used as template a 293-bp fragment from positions -213 to +80 relative to its TSP (Ainsa et al., 1999), and MtrA protected a 40-nucleotide region, from positions -68 to -29, that overlaps the -35 sequence of the whiI promoter (Figure 5C). To test the ability of MtrA to bind the protected sequence of bldD in EMSAs, a short oligonucleotide containing this region was used as a probe and was shifted completely by MtrA even with the lowest amount of protein tested (Figure 6A). Similarly, MtrA also retarded completely a short probe containing the protected sequence upstream of whiI (Figure 6A).
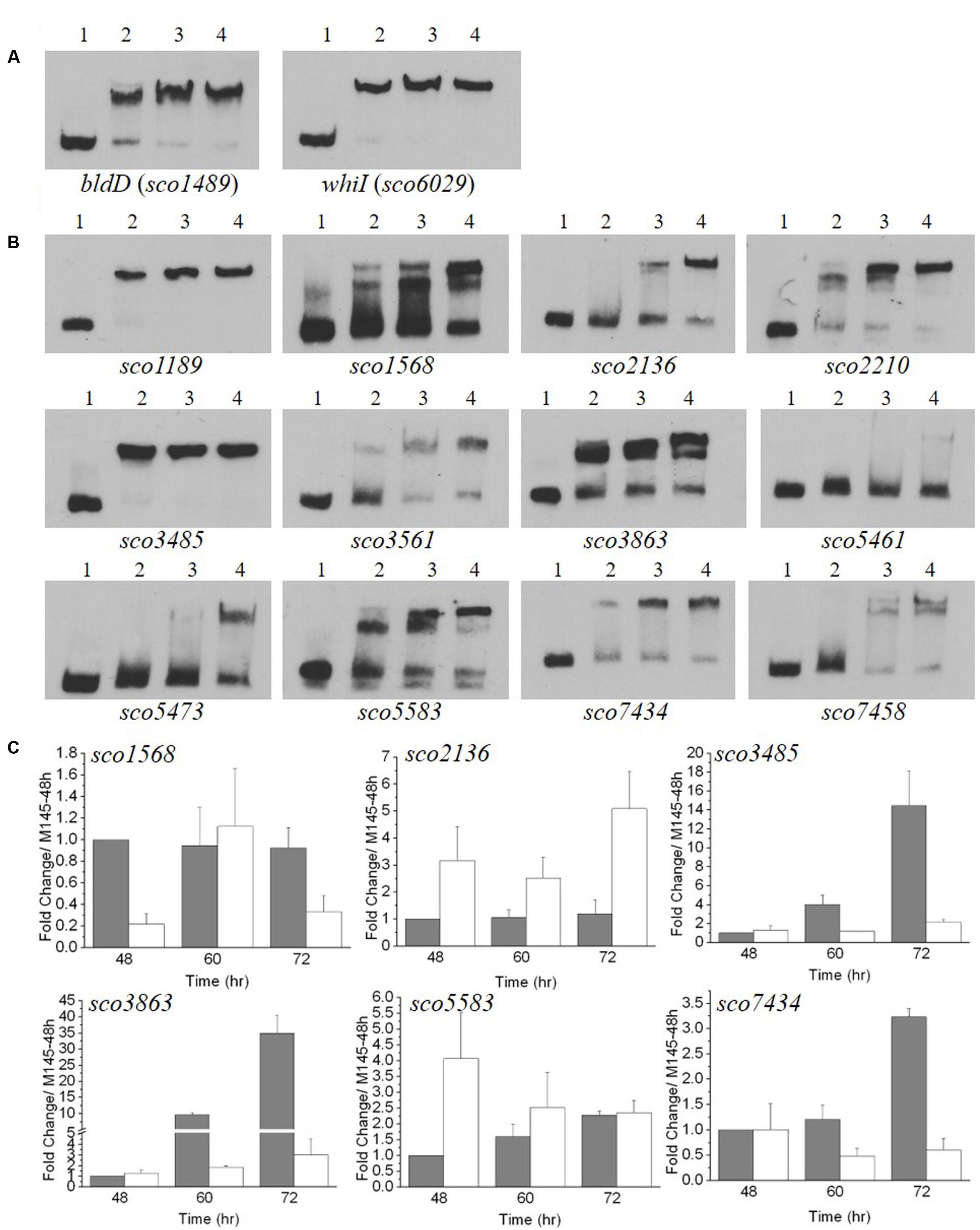
FIGURE 6. Electrophoretic mobility shift assays (EMSA) and transcriptional analysis of genes with upstream sequences similar to the MtrAMTB recognition sequence. (A,B) EMSA analyses with MtrASCO and 59-bp oligonucleotides containing sequences similar to the MtrAMTB recognition sequence. A fixed amount of labeled oligonucleotide was incubated in reactions containing no MtrA (lane 1), or 1.6 or 3.2, or 4.8 μg MtrA (lanes 2, 3, and 4, respectively). (A) Analysis with sequences upstream of bldD and whiI that were protected by MtrASCO in DNase I footprinting (see Figure 5). (B) Analysis with sequences identified in a genome-wide search for matches to the MtrAMTB consensus recognition sequence. The sequences were located upstream of the named genes. (C) Transcriptional analysis of potential MtrASCO target genes in the ΔmtrA mutant. M145 and ΔmtrA were grown on YBP solid medium, and RNA samples were isolated at the indicated times. Expression of hrdB, encoding the major sigma factor in M145, was used as an internal control. The y-axis shows the fold change in expression level in M145 (gray bars) over the level of ΔmtrA (light bars) at each time point, which was arbitrarily set to one. Results are the means (± SD) of triplet experiments.
MtrASCO has 75% amino acid identity to MtrAMTB and 69% amino acid identity to MtrACGL (Hoskisson and Hutchings, 2006). The consensus sequences recognized by MtrAMTB and MtrACGL have been characterized in vitro (Brocker et al., 2011) and in vivo (Galagan et al., 2013), respectively, revealing a similar, loosely conserved sequence composed of two 6-bp direct repeats (Supplementary Figure S4). These repeats are biased toward A/T for MtrACGL and toward G/C for MtrAMTB (Brocker et al., 2011; Galagan et al., 2013), possibly owing to the lower G+C content of the genome of C. glutamicum compared to that of M. tuberculosis (Cole et al., 1998; Kalinowski et al., 2003). Because of the higher sequence identity of MtrASCO to MtrAMTB and the similar G+C content of their genomes, we hypothesized that the consensus sequence recognized by MtrASCO would more closely resemble the one recognized by MtrAMTB. An examination of the sequences protected by MtrASCO upstream of bldD (Figure 5B) and whiI (Figure 5C) revealed matches to the MtrAMTB consensus, suggesting the two proteins have similar binding motifs.
The genome of S. coelicolor M145 was searched using the MtrAMTB binding consensus sequence (Galagan et al., 2013), and multiple potential recognition sites for MtrASCO were discovered (data not shown). The sites with high scores were selected, and the ability of MtrASCO to bind these sites was tested using short, oligonucleotide probes. In EMSA analysis, almost complete shifting was observed with probes for regions upstream of sco1189 and sco3485, with the least amount of MtrA protein tested (Figure 6B). With increasing amounts of MtrA, increased shifting was detected with probes for regions upstream of sco1568, sco2136, sco2210, sco3561, sco3863, sco5583, sco7434, and sco7458. However, probes for regions upstream of sco5461 and sco5473 were shifted only with the highest amounts of MtrA (Figure 6B). To test whether MtrA regulates these genes, transcription of the six genes was evaluated at 48, 60, and 72 h, with the transcription of the gene in M145 at 48 h arbitrarily set to one (Figure 6C). Expression of four genes, sco1568, sco3485, sco3863, and sco7434, was significantly down-regulated in ΔmtrA in at least one time point (Figure 6C), whereas sco2136 and sco5583 were up-regulated in ΔmtrA in at least one time point (Figure 6C), confirming that MtrA regulates these genes and consistent with the EMSA data.
Mutational Analyses of the MtrASCO Consensus Binding Site
To determine a consensus binding sequence for MtrASCO, the 13 probes, including those for bldD and whiI, that had stronger affinity to MtrASCO in EMSA analysis (Figures 6A,B) were used as input sequences for MEME analysis (Figure 7A). An 18-nucleotide consensus sequence with two, imperfect direct repeats of five nucleotides was revealed for MtrASCO binding (Figure 7B), a sequence very similar to the consensus binding sequences for MtrAMTB and MtrACGL, although with some variation in the degree of conservation for each nucleotide (Supplementary Figure S4). To validate the role of the consensus sequence in MtrASCO binding, we performed EMSAs using oligonucleotide probes containing point mutations in the repeat motifs upstream of bldD (Figure 7C). Close analysis of the sequence had revealed two overlapping sets of repeats that shared one half-site, and in contrast to the other sites (Figure 7A), the repeats in bldD-a are separated by one less nucleotide. Mutation of as few as three nucleotides in the central half-site markedly decreased MtrA binding in comparison to that observed with the native sequence (Figure 7D). Mutation of the three conserved nucleotides in the rightmost half-site only slightly reduced MtrA binding (Supplementary Figure S5), whereas mutation of three of the four conserved nucleotides in the left most motif markedly reduced MtrA binding (Figure 7E), suggesting that the left set of repeats may be more important for MtrA binding to the bldD promoter.
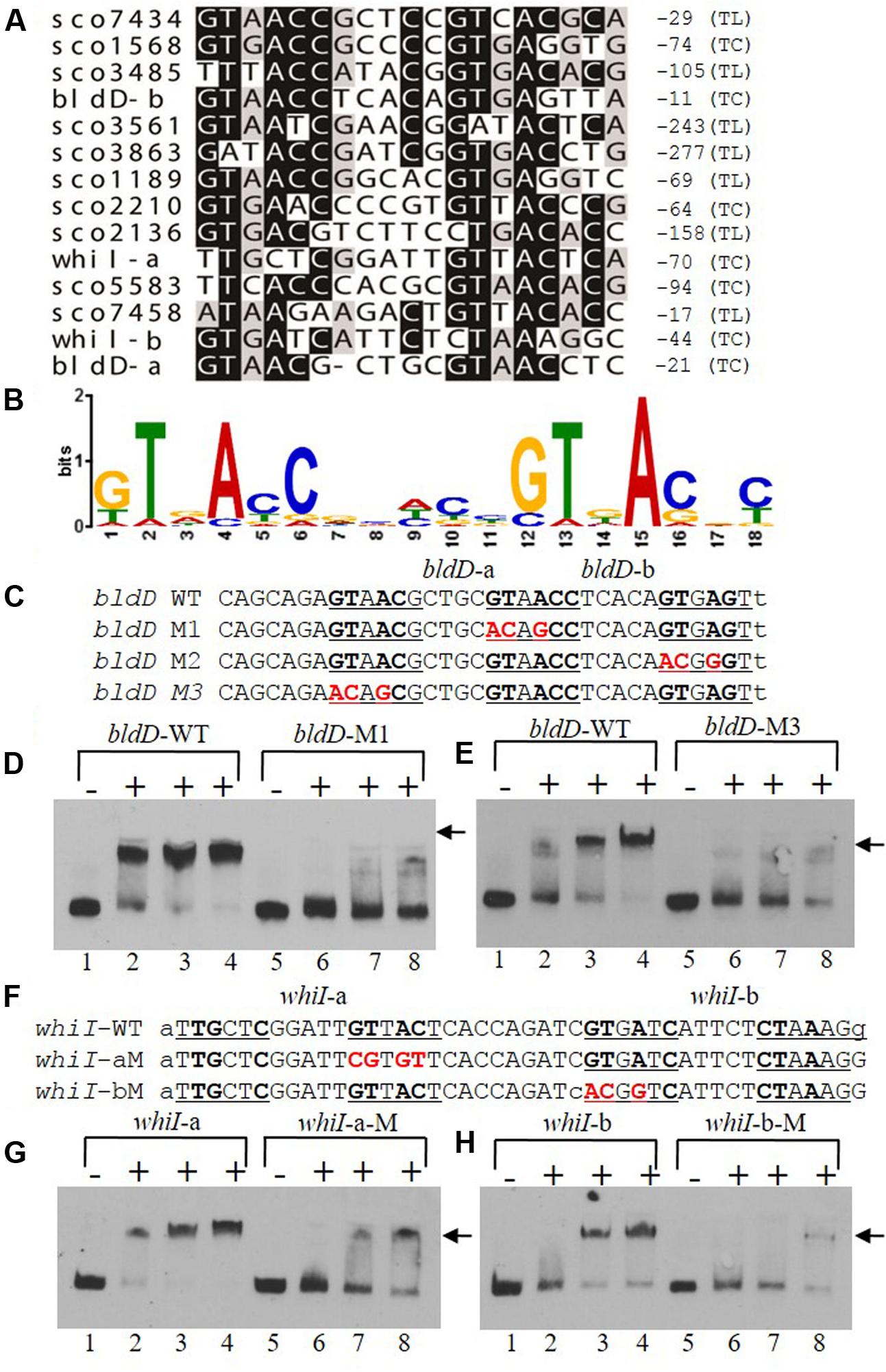
FIGURE 7. A consensus binding sequence for MtrASCO and mutational analysis of predicted binding sites in the bldD and whiI promoters. (A) Alignment of MtrA-shifted sequences (see Figure 6), with conserved nucleotides indicated by a darkened background. Numbers at the right indicate distance from putative translational start site (TL) or transcriptional start site (TC). Two putative sites are present upstream of bldD (bldD-a and bldD-b) and whiI (whiI-a and whiI-b). (B) Consensus sequence for MtrA recognition based on alignment in (A) and containing two direct repeats of approximately six nucleotides. (C–H) Mutational analyses of putative MtrA binding sites in the promoters of (C–E) bldD and (F–H) whiI. Mutagenized bases are shown in red, and probes with these mutations were compared with the wild-type sequence for binding to MtrA in EMSAs. (D,E,G,H) Reactions were carried out with the addition of no MtrA (lanes 1, 5); 2.1 μg MtrA (lanes 2, 6); 4.2 μg MtrA (lanes 3, 7); or 6.3 μg MtrA (lanes 4, 8). Arrowheads indicate the positions of the shifted probes.
Two potential MtrA binding sites were identified upstream of whiI in the MtrA-protected sequence (Figure 7F). The first site, designated whiI-a, includes positions -69 to -53 and the second site, designated whiI-b, includes positions -43 to -27, relative to the TSP of whiI (Figure 7F). EMSAs suggested that MtrA has stronger affinity for whiI-a than for whiI-b (Figures 7G,H), possibly because whiI-a has more conserved nucleotides in the direct repeats than does whiI-b (Figures 7A,F). Mutation of three or four of the conserved nucleotides within whiI-a or whiI-b probes severely reduced MtrA binding (Figures 7G,H). These data suggest that the whiI promoter contains more than one MtrA binding site and that the conserved nucleotides contribute to MtrA binding. Collectively, these data support the consensus sequence identified for MtrA binding.
Mutation of the mtrA Homologs in S. venezuelae and S. lividans Leads to a Conditional Bald Phenotype
BLAST analysis indicated that proteins with high similarity to MtrASCO are present in other Streptomyces species, including S. griseus, S. venezuelae, S. clavuligerus, S. scabies, and S. avermitilis1, reaching 98–100% amino acid identity. To investigate the functional conservation of MtrA in other Streptomyces species, the coding sequence of sve_2756, the mtrA homolog in S. venezuelae, was deleted in the wild-type strain ISP5320, generating Δsve_2756. Δsve_2756 displayed a bald phenotype, especially on rich medium, with significantly delayed formation of aerial mycelium on R5, R2YE, and YBP media (Figure 8A). On YBP solid medium, growth of aerial mycelium was observed as early as 48 h for ISP5320; however, Δsve_2756 remained bald even after 8 days of growth under the same conditions (Figure 8A). The complemented strain C-Δsve_2756 was generated using pCom-2756SV E, and as expected, this complementation restored the morphological defect (Figure 8A), confirming that the bald phenotype of Δsve_2756 is a direct result of sve_2756 mutation and indicating that SVE_2756 is required for normal development in S. venezuelae. However, some mutant strains of S. venezuelae appear bald, but in fact produce precocious hypersporulation rather than fail to form aerial hyphae (Tschowri et al., 2014). To determine whether the bald phenotype of Δsve_2756 was due to defective formation of aerial mycelium, its morphology was observed by SEM (Figure 8B), using strains grown for 84 h on YBP solid medium. Both the wild-type ISP5320 and the complemented strain C-Δsve_2756 were abundantly covered with straight spore chains, whereas Δsve_2756 was covered only by vegetative mycelia (Figure 8B), confirming that Δsve_2756 is defective in the formation of the aerial mycelium, and indicating an essential role for SVE_2756 in normal morphological development in S. venezuelae.
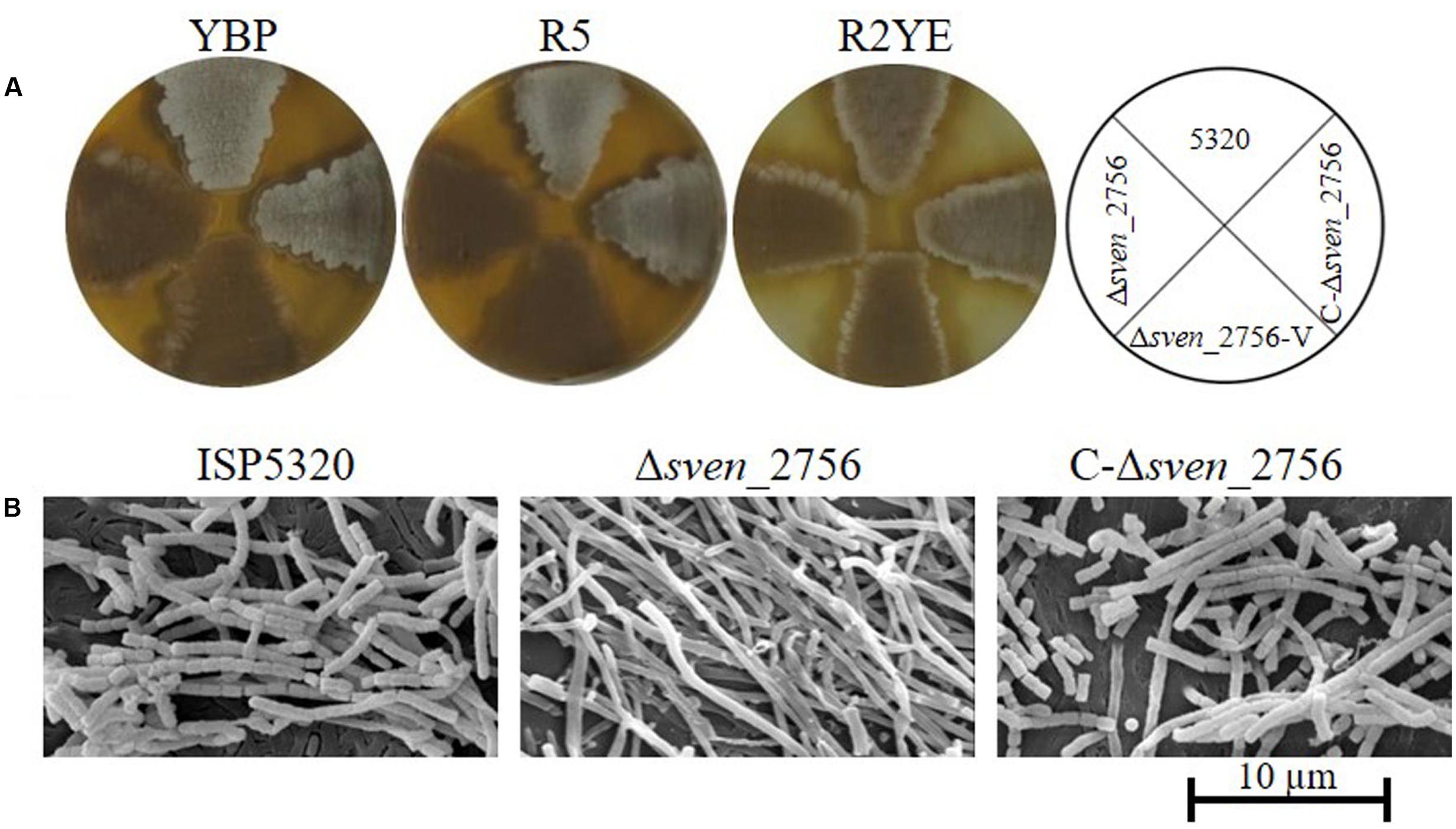
FIGURE 8. Requirement of the mtrA homolog sven_2756 for the formation of aerial hyphae in S. venezuelae. (A) Phenotypes of the S. venezuelae wild-type strain ISP5320, the deletion mutant Δsven_2756, and the sven_2756-complemented strain C-Δsve_2756 grown at 30°C on solid YBP (5 days), R5 (5 days), and R2YE media (84 h). (B) SEM images of S. venezuelae ISP5320, Δsven_2756, and C-Δsven_2756 strains after growth on YBP agar for 84 h. Images reveal the classic ‘bald’ phenotype for the deletion mutant and normal aerial hyphae for the wild-type and complemented strains. The scale bar is 10 μm.
Streptomyces lividans is nearly identical to S. coelicolor in sequence, but has obvious differences in morphology and physiology (Lewis et al., 2010). To investigate the role of the mtrA homolog sli_3357 in S. lividans, the deletion mutant Δsli_3357 was generated in strain 1326. Strain Δsli_3357 exhibited slightly delayed formation of aerial mycelium on MS medium (Supplementary Figure S6A), with a more obvious delay on R2YE and YBP media (Supplementary Figure S6A). In the complemented strain C-Δsli_3357, generated using pCom-3013SCO, growth of aerial mycelium was restored to levels comparable to that of the parental strain 1326 (Supplementary Figure S6A). SEM of cultures grown on YBP solid medium revealed that the wild-type strain 1326 and complemented strain C-Δsli_3357 were covered with abundant, straight or slightly coiled spore chains, whereas only fragments of submerged mycelium were observed for Δsli_3357 (Supplementary Figure S6B), confirming that Δsli_3357 is defective in the growth of aerial mycelia and indicating SLI_3357 is required for development in S. lividans. Altogether, these results suggest that MtrA homologs have a similar and essential role in the normal development of Streptomyces species.
Discussion
MtrA Is a New Member of Developmental Regulators in Streptomcyes
Our study showed that mutation of the response regulator gene mtrA leads to the bald phenotype characteristic of bld mutants, suggesting that MtrA is a new developmental regulator in S. coelicolor. We further determined that MtrA controls multiple genes critical for development, most notably bldD, bldK, whiH, and whiI; additionally, bldD and whiI were characterized as MtrA targets through in vitro analysis. As transcription of chp, rdl, and ram genes was also markedly influenced by deletion of mtrA in our study, we investigated whether MtrA regulates these genes directly or indirectly. Our preliminary data showed that MtrASCO interacts with the upstream sequence of several chp, rdl, and ram genes (Supplementary Figure S7), implying potentially direct control of MprASCO over these structural components of aerial mycelium. Although the data in this study were obtained mostly from in vitro analysis, such as EMSAs, our data are consistent with a recent study carried out in S. venezuelae that characterized the in vivo targets of MprASV E using the Chip-Seq assay (Som et al., 2017). Som et al. (2017) reported that whiI, which we identified as a direct target of MtrASCO, was one of the four in vivo MtrA targets with a statistically significant high enrichment value in S. venezuelae using Chip-Seq. Moreover, several bld genes (bldG, bldH, bldM, and bldN), whi genes (whiB, whiD, whiG, and whiH), chp genes (chpH, chpE, chpG, chpF, chpG, and chpD), rdlAB, and sapB displayed moderate to low levels of enrichment in Chip-Seq analysis, indicating that these genes are potential MtrA targets in S. venezuelae, consistent with many of our in vitro findings for S. coelicolor. However, bldD, which we found to be a direct target of MtrA through in vitro assays, was not identified as an MtrA target in S. venezuelae, possibly due to species differences.
Based on the analyses for MtrA in S. coelicolor and S. venezuelae, we propose a simple model for the MtrA regulatory network in governing development and differentiation in S. coelicolor (Figure 9). In this model, on receiving unknown environmental signals, the sensor kinase MtrB phosphorylates its cognate regulator MtrA, and the phosphorylated MtrA activates the chp and rdl genes that make up the components of aerial mycelium, and the ramCSAB operon at the early phase, resulting in the production of SapB and promoting the formation of aerial mycelium. MtrA also initiates, although possibly indirectly, the transcription of the bldK cluster, thus enabling transport of an early signal molecule into the cell (Nodwell et al., 1996), which also has a positive effect on SapB production and expression of chp and rdl genes through as yet unknown mechanisms. MtrA directly represses bldD, and the resulting lower levels of BldD allow activation of BldN whose targets include chp and rdl genes (Elliot et al., 2003; Bibb et al., 2012). The bald phenotype of ΔmtrA is thus likely a combined effect of the deeply blocked expression of chaplins and rodlins, which are the structural components of aerial mycelium; the minimal production level of SapB, a protein that assists in the erection of vegetative mycelium into the air; the blocked transcription of bldK, which results in the failed import of the early signal molecule critical for cellular development; and the overexpression of bldD, which may help maintain the mycelium in the vegetative phase. Additionally, MtrA also regulates whiI directly and whiH indirectly, and as expression of both genes is essential for activation of whiE (Kelemen et al., 1998), MtrA thus impacts Streptomyces differentiation through the whiIHE pathway (Figure 9).
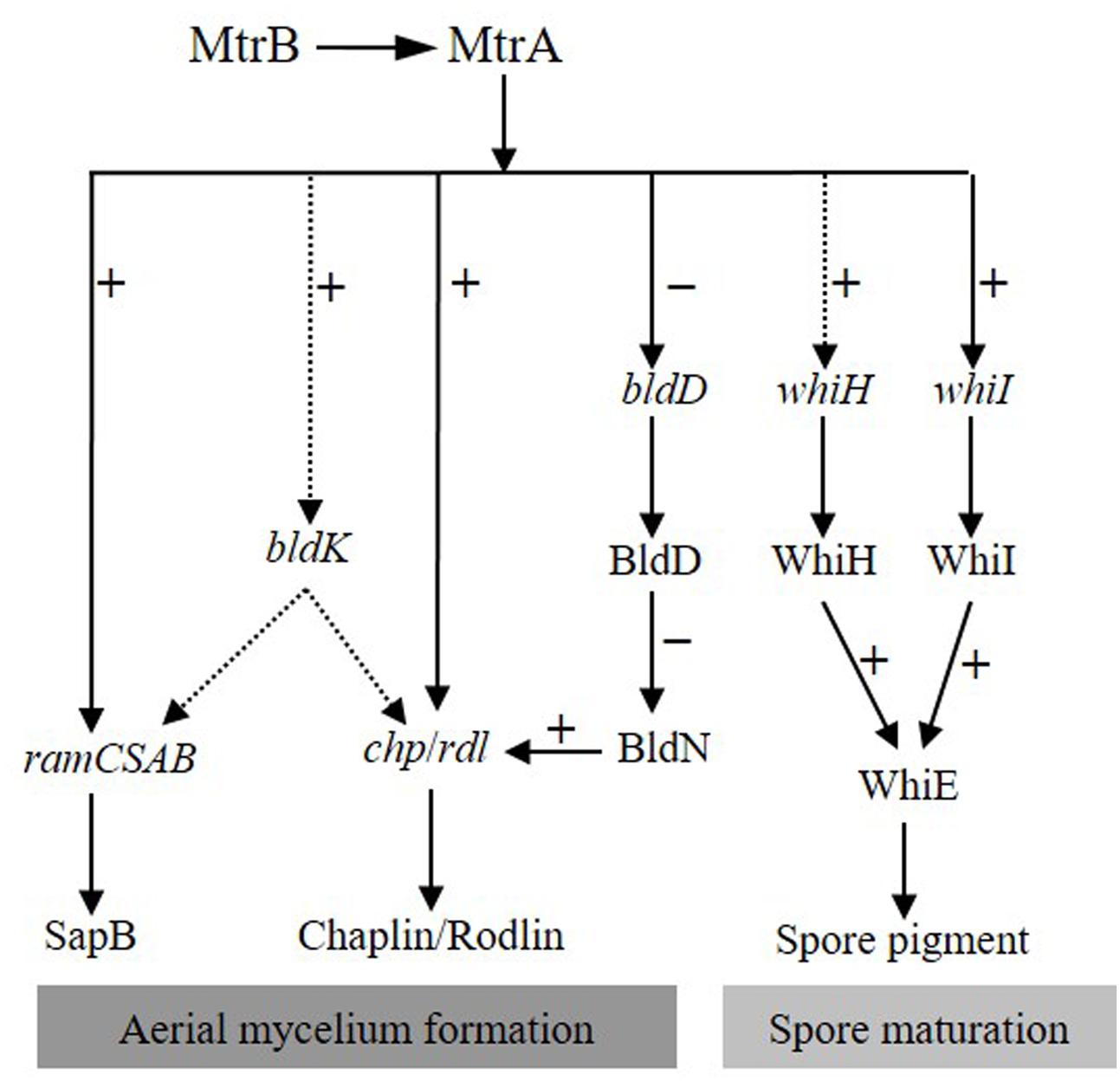
FIGURE 9. Proposed model of the MtrA regulatory network governing development and differentiation in S. coelicolor. In response to a currently unidentified environmental trigger, MtrA is activated by MtrB phosphorylation, and then the activated MtrA activates chp, rdl, and ram genes and the bldK cluster, the latter of which may also have a positive regulatory effect on the expression of ram, rdl, and chp genes. In contrast, MtrA represses bldD, allowing activation of bldN, a sigma factor gene repressed by BldD-c-di-GMP, and BldN then activates chp and rdl genes, further amplifying the effect of MtrA on the regulation of aerial mycelium formation. MtrA regulates the expression of whiI and whiH, both of which are required for expression of whiE, thus impacting differentiation through the whiIHE pathway. Solid and dotted arrows indicate direct and indirect regulation, respectively. Minus (–) and plus (+) signs indicate a negative or positive role in the regulation of the downstream gene.
MtrA Is a Transcriptional Regulator of bldD
Streptomyces development is a complex process controlled by bld and whi genes, which form a developmentally regulatory network. BldD seems to be a master regulator of this network, with the BldD-c-di-GMP complex directly controlling bldA, bldC, bldH, bldM, bldN, whiB, and whiG (Den Hengst et al., 2010), and indirectly controlling whiH, whiI, and whiE (Bush et al., 2015, 2016). BldD is autoregulatory, repressing its own expression (Elliot et al., 1998), but no other regulators of bldD were previously identified. We found that MtrA can bind the region upstream of bldD, that mutation of an MtrA consensus target sequence reduces MtrA binding to this region, and that deletion of mtrA results in enhanced expression of bldD, revealing MtrA as a second regulator of bldD. Previous studies have shown that, when wild-type S. coelicolor is grown on solid R5 or R2YE, two types of media with essentially the same components, bldD expression peaks at the very early growth phase (about 14 to 15 h), decreases by 24 h, although still remaining relatively high, and then becomes very low by 48 h (Elliot et al., 1998; Den Hengst et al., 2010). Consistent with these reports, we observed decreased bldD expression between 24 and 48 h in M145. However, the expression pattern of bldD after 48 h has not been previously reported, and unexpectedly, we detected increased expression at later time points compared to the level at 48 h. Of particular interest, the bldD expression level in ΔmtrA was consistently higher than in M145 at all the time points tested, with the highest expression levels detected in the mutant from 48 h to 84 h, suggesting that bldD is derepressed, especially at the late growth phase in ΔmtrA. As a global regulator, BldD represses genes essential for morphological development and sporulation (Elliot et al., 2001; Den Hengst et al., 2010; Tschowri et al., 2014), and it was proposed that BldD represses premature expression of bld target genes during vegetative growth (Elliot et al., 2001; Tschowri et al., 2014). Notably, although both BldD (Elliot et al., 1998) and MtrA repress bldD transcription, bldD was significantly derepressed at the early stage in a bldD mutant (Elliot et al., 1998), whereas derepression of bldD was most evident at the late phase in ΔmtrA, potentially reflecting different regulatory roles. Interestingly, the MtrA-protected sequence overlaps the BldD sites in the bldD promoter (Elliot et al., 2001; Den Hengst et al., 2010), suggesting that BldD and MtrA may interact to control this gene in S. coelicolor.
MtrASCO Has an Important but a Different Role, Compared with MtrAMTB and MtrACGL
MtrA has been primarily studied in M. tuberculosis and C. glutamicum, displaying differing but critical roles in these organisms (Zahrt and Deretic, 2000, 2001; Moker et al., 2004). In addition to its essential role in cell viability, MtrAMTB is also implicated in modulating cell proliferation (Fol et al., 2006), by sequestering oriC (Purushotham et al., 2015). The primary targets of MtrAMTB include dnaA; fbpB, which encodes the major secreted immunodominant antigen Ag85B; and rpfB, encoding the resuscitation-promoting factor (Li et al., 2010; Rajagopalan et al., 2010; Sharma et al., 2015). The contribution of MtrACGL is manifested in cell morphology, resistance to antibiotics, and osmoprotection (Moker et al., 2004), and its target genes have diverse functions, including genes encoding a putative cell wall peptidase, transporters, and membrane proteins (Moker et al., 2004; Brocker and Bott, 2006; Brocker et al., 2011). Our study revealed a unique function for MtrA in Streptomyces, where it has a major role in the developmental life cycle, controlling multiple genes involved in the formation of the aerial mycelium.
In a system-wide study to characterize the global regulatory network of transcription factors (TFs) in M. tuberculosis, target sequences for TFs were identified by Chip-seq analysis, and for MtrAMTB, a conserved recognition sequence composed of two imperfect direct repeats of six nucleotides was deduced from more than sixty target sequences (Galagan et al., 2013). A binding motif similar to that of MtrAMTB but more biased to A/T was deduced through in vitro analysis for MtrACGL (Brocker et al., 2011). We identified similar direct repeats of 5–6 nucleotides, G/T-T-G/A-A-C-C-NNNNN-G-T-G/T-A-C-N, separated by five nucleotides, in regions bound by MtrASCO, including a site that overlaps the -10 region in the bldD promoter, potentially explaining how MtrA represses bldD. For whiI, two potential MtrASCO sites, whiI-a and whiI-b, were found upstream of whiI, with whiI-b overlapping the -35 region and whiI-a upstream of the -35 region (from positions -68 to -53). Given that binding to the whiI-b site could potentially inhibit transcription, the positive regulatory effect of MtrA over whiI expression implies that whiI-a maybe the major binding site for MtrA, and notably, EMSAs analysis indicated that MtrA appears to have a higher affinity for whiI-a than for whiI-b. In addition, as noted above, our conclusion that whiI is a direct target of MtrA is further supported by the recent in vivo study on MtrA targets in S. venezuelae (Som et al., 2017). Also consistent with the S. venezuelae study, we detected interactions between MtrASCO and upstream regions of chp genes, rdl, and ramC, and found loosely conserved MtrA sites upstream of these genes (Supplementary Figure S7). Additionally, our RNA-Seq and real-time PCR analyses on the parental strain and ΔmtrA indicate that these genes are positively regulated by MtrA. Although transcription of whiB and whiG did not appear to be altered in ΔmtrA under the conditions used in our study (Supplementary Figure S3), potential MtrA sites were predicted upstream of these two genes (Supplementary Table S3), consistent with the in vivo study in S. venezuelae, which identified whiB and whiG as MtrA targets (Som et al., 2017).
Any Other Role(s) of MtrA in Streptomyces?
In addition to its role in development, are there any other functions for MtrA in Streptomyces? Our preliminary analyses indicate that production of the signature blue pigment actinorhodin in ΔmtrA differs from that in S. coelicolor M145 (Figure 1A), and we therefore hypothesize that, in addition to a key role in regulating development, MtrA also contributes to the regulation of secondary metabolism in S. coelicolor. Furthermore, a genome-wide search of S. coelicolor revealed 100s of intergenic sequences highly similar to the MtrASCO consensus binding sequence (Supplementary Table S3), suggesting that MtrA is a global regulator in S. coelicolor, although the function of many of these genes is not yet known. Additional studies on the intriguing roles of this TCS are ongoing.
Author Contributions
XP conceived and supervised the studies, and wrote the paper; PZ, LW, and YZ performed all experiments; ML carried out bioinformatics analysis; GC, YW, X-LC, MT, and XP analyzed the data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (NFSC31470165 to XP and NFSC31670038 to X-LC), by Open Funding Project of State Key Laboratory of Microbial Metabolite (MMLKF15-01 to XP), by the Innovation Project of Shandong Academy of Medical Sciences (201604 to GC) and by the State Key Laboratory of Microbial Technology. We thank Prof. Keith F. Chater for providing the PCR-targeting system.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02013/full#supplementary-material
Footnotes
References
Ainsa, J. A., Parry, H. D., and Chater, K. F. (1999). A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34, 607–619. doi: 10.1046/j.1365-2958.1999.01630.x
Al-Bassam, M. M., Bibb, M. J., Bush, M. J., Chandra, G., and Buttner, M. J. (2014). Response regulator heterodimer formation controls a key stage in Streptomyces development. PLOS Genet. 10:e1004554. doi: 10.1371/journal.pgen.1004554
Bailey, T. L., and Elkan, C. (1994). “Fitting a mixture model by expectation maximization to discover motifs in biopolymers,” in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, eds R. Altman, D. Brutlag, P. Karp, R. Lathrop, and D. Searls (Menlo Park, CA: AAAI Press), 28–36.
Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bibb, M. J., Domonkos, A., Chandra, G., and Buttner, M. J. (2012). Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by sigma(BldN) and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 84, 1033–1049. doi: 10.1111/j.1365-2958.2012.08070.x
Brocker, M., and Bott, M. (2006). Evidence for activator and repressor functions of the response regulator MtrA from Corynebacterium glutamicum. FEMS Microbiol. Lett. 264, 205–212. doi: 10.1111/j.1574-6968.2006.00456.x
Brocker, M., Mack, C., and Bott, M. (2011). Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J. Bacteriol. 193, 1237–1249. doi: 10.1128/JB.01032-10
Bush, M. J., Bibb, M. J., Chandra, G., Findlay, K. C., and Buttner, M. J. (2013). Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. MBio 4, e00684–13. doi: 10.1128/mBio.00684-13
Bush, M. J., Chandra, G., Bibb, M. J., Findlay, K. C., and Buttner, M. J. (2016). Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. MBio 7:e00523–16. doi: 10.1128/mBio.00523-16
Bush, M. J., Tschowri, N., Schlimpert, S., Flardh, K., and Buttner, M. J. (2015). c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 13, 749–760. doi: 10.1038/nrmicro3546
Cao, G., Howard, S. T., Zhang, P., Wang, X., Chen, X. L., Samten, B., et al. (2015). EspR, a regulator of the ESX-1 secretion system in Mycobacterium tuberculosis, is directly regulated by the two-component systems MprAB and PhoPR. Microbiology 161, 477–489. doi: 10.1099/mic.0.000023
Capstick, D. S., Willey, J. M., Buttner, M. J., and Elliot, M. A. (2007). SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol. Microbiol. 64, 602–613. doi: 10.1111/j.1365-2958.2007.05674.x
Chater, K. (2011). “Differentiation in Streptomyces: the properties and programming of diverse cell-types,” in Streptomyces: Molecular Biology and Biotechnology, ed. P. Dyson (Poole: Caister Academic Press), 43–86.
Claessen, D., Rink, R., De Jong, W., Siebring, J., De Vreugd, P., Boersma, F. G., et al. (2003). A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726. doi: 10.1101/gad.264303
Claessen, D., Stokroos, I., Deelstra, H. J., Penninga, N. A., Bormann, C., Salas, J. A., et al. (2004). The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53, 433–443. doi: 10.1111/j.1365-2958.2004.04143.x
Claessen, D., Wosten, H. A., Van Keulen, G., Faber, O. G., Alves, A. M., Meijer, W. G., et al. (2002). Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 44, 1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Davis, N. K., and Chater, K. F. (1990). Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol. Microbiol. 4, 1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x
de Jong, W., Wosten, H. A., Dijkhuizen, L., and Claessen, D. (2009). Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol. Microbiol. 73, 1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x
Den Hengst, C. D., Tran, N. T., Bibb, M. J., Chandra, G., Leskiw, B. K., and Buttner, M. J. (2010). Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78, 361–379. doi: 10.1111/j.1365-2958.2010.07338.x
Duong, A., Capstick, D. S., Di Berardo, C., Findlay, K. C., Hesketh, A., Hong, H. J., et al. (2012). Aerial development in Streptomyces coelicolor requires sortase activity. Mol. Microbiol. 83, 992–1005. doi: 10.1111/j.1365-2958.2012.07983.x
Elliot, M., Damji, F., Passantino, R., Chater, K., and Leskiw, B. (1998). The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 180, 1549–1555.
Elliot, M. A., Bibb, M. J., Buttner, M. J., and Leskiw, B. K. (2001). BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40, 257–269. doi: 10.1046/j.1365-2958.2001.02387.x
Elliot, M. A., Karoonuthaisiri, N., Huang, J., Bibb, M. J., Cohen, S. N., Kao, C. M., et al. (2003). The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17, 1727–1740. doi: 10.1101/gad.264403
Elliot, M. A., and Leskiw, B. K. (1999). The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J. Bacteriol. 181, 6832–6835.
Fink, D., Weissschuh, N., Reuther, J., Wohlleben, W., and Engels, A. (2002). Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 46, 331–347. doi: 10.1046/j.1365-2958.2002.03150.x
Flardh, K., and Buttner, M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49. doi: 10.1038/nrmicro1968
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., et al. (2006). Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657. doi: 10.1111/j.1365-2958.2006.05137.x
Galagan, J. E., Minch, K., Peterson, M., Lyubetskaya, A., Azizi, E., Sweet, L., et al. (2013). The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183. doi: 10.1038/nature12337
Gaskell, A. A., Giovinazzo, J. A., Fonte, V., and Willey, J. M. (2012). Multi-tier regulation of the streptomycete morphogenetic peptide SapB. Mol. Microbiol. 84, 501–515. doi: 10.1111/j.1365-2958.2012.08041.x
Gregory, M. A., Till, R., and Smith, M. C. (2003). Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J. Bacteriol. 185, 5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003
Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546. doi: 10.1073/pnas.0337542100
Hoskisson, P. A., and Hutchings, M. I. (2006). MtrAB-LpqB: a conserved three-component system in actinobacteria? Trends Microbiol. 14, 444–449.
Hutchings, M. I., Hoskisson, P. A., Chandra, G., and Buttner, M. J. (2004). Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150, 2795–2806. doi: 10.1099/mic.0.27181-0
Kalinowski, J., Bathe, B., Bartels, D., Bischoff, N., Bott, M., Burkovski, A., et al. (2003). The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104, 5–25. doi: 10.1016/S0168-1656(03)00154-8
Keijser, B. J., Van Wezel, G. P., Canters, G. W., and Vijgenboom, E. (2002). Developmental regulation of the Streptomyces lividans ram genes: involvement of RamR in regulation of the ramCSAB operon. J. Bacteriol. 184, 4420–4429. doi: 10.1128/JB.184.16.4420-4429.2002
Kelemen, G. H., Brian, P., Flardh, K., Chamberlin, L., Chater, K. F., and Buttner, M. J. (1998). Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3 (2). J. Bacteriol. 180, 2515–2521.
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (eds) (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kodani, S., Hudson, M. E., Durrant, M. C., Buttner, M. J., Nodwell, J. R., and Willey, J. M. (2004). The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 101, 11448–11453. doi: 10.1073/pnas.0404220101
Lewis, R. A., Laing, E., Allenby, N., Bucca, G., Brenner, V., Harrison, M., et al. (2010). Metabolic and evolutionary insights into the closely-related species Streptomyces coelicolor and Streptomyces lividans deduced from high-resolution comparative genomic hybridization. BMC Genomics 11:682. doi: 10.1186/1471-2164-11-682
Li, Y., Zeng, J., Zhang, H., and He, Z. G. (2010). The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol. 10:242. doi: 10.1186/1471-2180-10-242
Ma, H., and Kendall, K. (1994). Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176, 3800–3811. doi: 10.1128/jb.176.12.3800-3811.1994
McCormick, J. R., and Flardh, K. (2011). Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36, 206–231. doi: 10.1111/j.1574-6976.2011.00317.x
Moker, N., Brocker, M., Schaffer, S., Kramer, R., Morbach, S., and Bott, M. (2004). Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54, 420–438. doi: 10.1111/j.1365-2958.2004.04249.x
Molle, V., and Buttner, M. J. (2000). Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36, 1265–1278. doi: 10.1046/j.1365-2958.2000.01977.x
Nguyen, K. T., Willey, J. M., Nguyen, L. D., Nguyen, L. T., Viollier, P. H., and Thompson, C. J. (2002). A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 46, 1223–1238. doi: 10.1046/j.1365-2958.2002.03255.x
Nodwell, J. R., Mcgovern, K., and Losick, R. (1996). An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22, 881–893. doi: 10.1046/j.1365-2958.1996.01540.x
O’Connor, T. J., Kanellis, P., and Nodwell, J. R. (2002). The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45, 45–57. doi: 10.1046/j.1365-2958.2002.03004.x
Ou, X., Zhang, B., Zhang, L., Zhao, G., and Ding, X. (2009). Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl. Environ. Microbiol. 75, 2158–2165. doi: 10.1128/AEM.02209-08
Pospiech, A., and Neumann, B. (1995). A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11, 217–218. doi: 10.1016/S0168-9525(00)89052-6
Purushotham, G., Sarva, K. B., Blaszczyk, E., Rajagopalan, M., and Madiraju, M. V. (2015). Mycobacterium tuberculosis oriC sequestration by MtrA response regulator. Mol. Microbiol. 98, 586–604. doi: 10.1111/mmi.13144
Rajagopalan, M., Dziedzic, R., Al Zayer, M., Stankowska, D., Ouimet, M. C., Bastedo, D. P., et al. (2010). Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J. Biol. Chem. 285, 15816–15827. doi: 10.1074/jbc.M109.040097
Ryding, N. J., Kelemen, G. H., Whatling, C. A., Flardh, K., Buttner, M. J., and Chater, K. F. (1998). A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29, 343–357. doi: 10.1046/j.1365-2958.1998.00939.x
Sambrook, J., and Russell, D. W. (eds) (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Sharma, A. K., Chatterjee, A., Gupta, S., Banerjee, R., Mandal, S., Mukhopadhyay, J., et al. (2015). MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology 161, 1271–1281. doi: 10.1099/mic.0.000087
Sola-Landa, A., Moura, R. S., and Martin, J. F. (2003). The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U.S.A. 100, 6133–6138. doi: 10.1073/pnas.0931429100
Sola-Landa, A., Rodriguez-Garcia, A., Franco-Dominguez, E., and Martin, J. F. (2005). Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56, 1373–1385. doi: 10.1111/j.1365-2958.2005.04631.x
Som, N. F., Heine, D., Holmes, N. A., Munnoch, J. T., Chandra, G., Seipke, R. F., et al. (2017). The conserved actinobacterial two-component system MtrAB coordinates chloramphenicol production with sporulation in Streptomyces venezuelae NRRL B-65442. Front. Microbiol. 8:1145. doi: 10.3389/fmicb.2017.01145
Talbot, N. J. (2003). Aerial morphogenesis: enter the chaplins. Curr. Biol. 13, R696–R698. doi: 10.1016/j.cub.2003.08.040
Tillotson, R. D., Wosten, H. A., Richter, M., and Willey, J. M. (1998). A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30, 595–602. doi: 10.1046/j.1365-2958.1998.01093.x
Tschowri, N., Schumacher, M. A., Schlimpert, S., Chinnam, N. B., Findlay, K. C., Brennan, R. G., et al. (2014). Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147. doi: 10.1016/j.cell.2014.07.022
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
Wildermuth, H. (1970). Surface structure of streptomycete spores as revealed by negative staining and freeze-etching. J. Bacteriol. 101, 318–322.
Wildermuth, H., Wehrli, E., and Horne, R. W. (1971). The surface structure of spores and aerial mycelium in Streptomyces coelicolor. J. Ultrastruct. Res. 35, 168–180. doi: 10.1016/S0022-5320(71)80149-1
Willey, J., Santamaria, R., Guijarro, J., Geistlich, M., and Losick, R. (1991). Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65, 641–650. doi: 10.1016/0092-8674(91)90096-H
Willey, J., Schwedock, J., and Losick, R. (1993). Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7, 895–903. doi: 10.1101/gad.7.5.895
Willey, J. M., Willems, A., Kodani, S., and Nodwell, J. R. (2006). Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 59, 731–742. doi: 10.1111/j.1365-2958.2005.05018.x
Wosten, H. A., and Willey, J. M. (2000). Surface-active proteins enable microbial aerial hyphae to grow into the air. Microbiology 146(Pt 4), 767–773. doi: 10.1099/00221287-146-4-767
Xu, Z., Wang, Y., Chater, K. F., Ou, H. Y., Xu, H. H., Deng, Z., et al. (2017). Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl. Environ. Microbiol. 83:e02889–16. doi: 10.1128/AEM.02889-16
Zahrt, T. C., and Deretic, V. (2000). An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182, 3832–3838. doi: 10.1128/JB.182.13.3832-3838.2000
Zahrt, T. C., and Deretic, V. (2001). Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. U.S.A. 98, 12706–12711. doi: 10.1073/pnas.221272198
Keywords: Streptomyces, MtrA, development, bld, whi
Citation: Zhang P, Wu L, Zhu Y, Liu M, Wang Y, Cao G, Chen X-L, Tao M and Pang X (2017) Deletion of MtrA Inhibits Cellular Development of Streptomyces coelicolor and Alters Expression of Developmental Regulatory Genes. Front. Microbiol. 8:2013. doi: 10.3389/fmicb.2017.02013
Received: 23 July 2017; Accepted: 29 September 2017;
Published: 16 October 2017.
Edited by:
Marc Bramkamp, Ludwig Maximilian University of Munich, GermanyReviewed by:
Matt Bush, John Innes Centre (BBSRC), United KingdomYuqing Li, Sichuan University, China
Copyright © 2017 Zhang, Wu, Zhu, Liu, Wang, Cao, Chen, Tao and Pang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuhua Pang, pangxiuhua@sdu.edu.cn Guangxiang Cao, caozhong0402@163.com
†These authors have contributed equally to this work.
 Peipei Zhang1†
Peipei Zhang1† Lili Wu
Lili Wu Guangxiang Cao
Guangxiang Cao Xiu-Lan Chen
Xiu-Lan Chen Xiuhua Pang
Xiuhua Pang