- 1School of Public Health, Zunyi Medical University, Zunyi, China
- 2Experimental Teaching Demonstration Center for Preventive Medicine of Guizhou Province, Zunyi Medical University, Zunyi, China
- 3Department of Pharmacology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 4Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
- 5Department of Food Science and Engineering, School of Science and Engineering, Jinan University, Guangzhou, China
- 6Department of Food Quality and Safety, Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences, Beijing, China
- 7Department of Food and Bioengineering, Beijing Agricultural Vocational College, Beijing, China
- 8China National Center for Food Safety Risk Assessment, Beijing, China
Ochratoxin A (OTA) is a toxic secondary fungal metabolite that widely takes place in various kinds of foodstuffs and feeds. Human beings and animals are inevitably threatened by OTA as a result. Therefore, it is necessary to adopt various measures to detoxify OTA-contaminated foods and feeds. Biological detoxification methods, with better safety, flavor, nutritional quality, organoleptic properties, availability, and cost-effectiveness, are more promising than physical and chemical detoxification methods. The state-of-the-art research advances of OTA biodetoxification by degradation, adsorption, or enzymes are reviewed in the present paper. Researchers have discovered a good deal of microorganisms that could degrade and/or adsorb OTA, including actinobacteria, bacteria, filamentous fungi, and yeast. The degradation of OTA to non-toxic or less toxic OTα via the hydrolysis of the amide bond is the most important OTA biodegradation mechanism. The most important influence factor of OTA adsorption capacity of microorganisms is cell wall components. A large number of microorganisms with good OTA degradation and/or adsorption ability, as well as some OTA degradation enzymes isolated or cloned from microorganisms and animal pancreas, have great application prospects in food and feed industries.
Introduction
Ochratoxin A (OTA), 7-carboxyl-5-chloro-8-hydroxyl-3,4-dihydro-3R-methyl-isocoumarin-7-L-β-phenylalanine (Figure 1; Wu et al., 2011), is a secondary fungal metabolite with low molecular weight that is mainly produced by various species of Aspergillus and Penicillium (Shen et al., 2013; Susca et al., 2016; Freire et al., 2017). Critical steps of OTA biosynthesis are shown in Figure 1 (Huff and Hamilton, 1979; Harris and Mantle, 2001; El Khoury and Atoui, 2010; Gallo et al., 2013, 2017; Wang et al., 2016). Common OTA-contaminated feeds (Streit et al., 2013; Li et al., 2014; Sherazi et al., 2015; Pinotti et al., 2016) and food commodities include cereals (maize, wheat, rice, sorghum, barley, oats, and rye) (Duarte et al., 2010; Liang et al., 2015; Lim et al., 2015; Sun et al., 2017), cereal products (bread, flour, and pasta) (Shen et al., 2014), wine (Quintela et al., 2013; Freire et al., 2017), dairy products (milk and cheese), meat (pork, fish, and poultry), eggs (Dai et al., 2014), fruit (grapes, apples, peaches, pears, strawberries, oranges, watermelons, mangoes, and figs) (Marin and Taranu, 2015; Freire et al., 2017), vegetables (yam, garlic, onions, potatoes, and tomatoes) (Pfohl-Leszkowicz and Manderville, 2007; Zhao et al., 2017), beans (coffee beans, peanuts, chickpeas, and soybeans) (Pfohl-Leszkowicz and Manderville, 2012; Wang et al., 2017), dried products (dried beans, smoked or salted dried fish, raisins, and jerky), nuts, sesame seeds, rapeseed, spice (Abrunhosa et al., 2010; Bui-Klimke and Wu, 2015), infant cereals (Cappozzo et al., 2017), and even milk-based baby formulae (Raiola et al., 2015; Zhao et al., 2017). According to the latest statistics from 2006 to 2016, the maximum concentration and incidence of OTA in raw cereal grains was 1,164 μg/kg and 29%, respectively (Lee and Ryu, 2017). Therefore, it is difficult to completely avoid the risk of OTA exposure. In 1993, OTA was classified as a group 2B carcinogen (possible human carcinogen) by the International Agency for Research on Cancer (IARC) (IARC, 1993). Although OTA has not been classified as a group 1 carcinogen (human carcinogen), such as aflatoxins, this does not mean that OTA is significantly less toxic than aflatoxins. In fact, OTA is a powerful carcinogen for rodents and poultry (Bondy et al., 2015), but the epidemiological evidence in human beings is very scarce (Bui-Klimke and Wu, 2015). That is to say, the carcinogenic evidence of OTA is not as sufficient as that of aflatoxins in human beings (Bui-Klimke and Wu, 2015). A great deal of animal or cell experiments have reported that the exposure of OTA can result in various toxicological effects, including the disruption of the gut microbiota homeostasis (Liew and Mohd-Redzwan, 2018), teratogenicity, carcinogenicity (Pfohl-Leszkowicz and Manderville, 2012), mutagenicity, hepatotoxicity (Zheng et al., 2013; Qi et al., 2014), genotoxicity (Pfohl-Leszkowicz and Manderville, 2007), immunotoxicity (Marin and Taranu, 2015), embryotoxicity (Hong et al., 2000), developmental toxicity, neurotoxicity (Bhat et al., 2016), testicular toxicity (Schwartz, 2002), blood-brain barrier damage (Jackson and Ryu, 2017), and especially nephrotoxicity (Zhao et al., 2017). The primary target organ of OTA is the kidney (Zhang et al., 2014). Although the human epidemiological evidence is inadequate, the association between OTA exposure and Balkan Endemic Nephropathy (BEN), Chronic Interstitial Nephropathy (CIN), and other kidney diseases is more or less existed (Bui-Klimke and Wu, 2015; Zhao et al., 2017). Thus, OTA attracts global concern based on its ubiquitous nature in feeds and foods and the adverse health effects in humans and animals (Duarte et al., 2010).
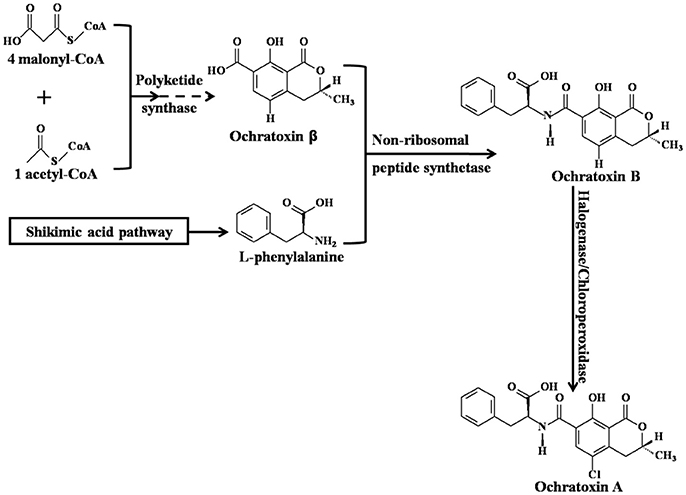
Figure 1. Critical steps of Ochratoxin A biosynthesis [adapted from (Gallo et al., 2017)].
Apart from causing health issues, OTA exposure results in enormous economic losses via decreasing the productivity of livestock and farm crops and increasing the medical fees of humans and animals, the mortality rate of animals, and the costs related to OTA precaution, control, and detoxication (Duarte et al., 2010; Pfliegler et al., 2015). The best way to avoid these negative health and economic effects would be preventing the contamination of OTA. But in fact, it is nearly impossible to completely avoid its contamination in feeds and foodstuffs. Instead, to adopt various measures to detoxify OTA-contaminated foods and feeds is more feasible and necessary (Russo et al., 2016). According to present reports, physical, chemical, and biological detoxification methods were mainly applied to food and feed industries (Kabak and Dobson, 2009; Quintela et al., 2013). Among these three classes of methods, biological methods, with better safety, flavor, nutritional quality, organoleptic properties, availability, and cost-effectiveness, were more promising than the other two classes (Kabak and Dobson, 2009; Rodriguez et al., 2011; Hathout and Aly, 2014; De Bellis et al., 2015; Cho et al., 2016; Farbo et al., 2016). Thus, this review principally focuses on the advances in biodetoxification of OTA.
Biodetoxification of OTA by Degradation
Microorganisms, including actinobacteria, bacteria, filamentous fungi, and yeast, that are able to degrade OTA are summarized in Table S1. Note that the direct in vivo experiments with animals and the in vitro experiments with animal tissues or body fluids that do not have specific microorganisms separated are not summarized in Tables S1, S2, such as the research of Xiao et al. (1991); Madhyastha et al. (1992); Özpinar et al. (1999); Müller et al. (1998), and Müller et al. (2001). In addition, microorganisms that could detoxify OTA but without explicit adsorption and/or biodegradation mechanisms of OTA elimination were neither shown in Tables S1, S2, such as the research of Böhm et al. (2000); Štyriak et al. (2001), and Caridi et al. (2006).
OTA Biodegradation Products and Their Properties
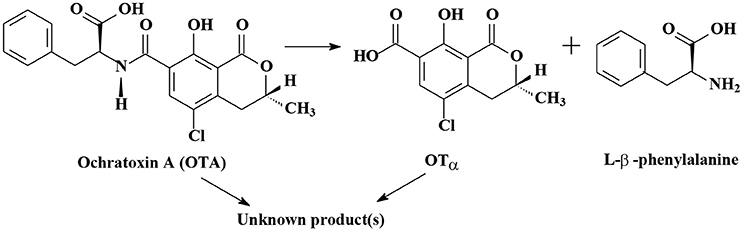
As shown in Table S1, the degradation of OTA to OTα (7-carboxy-5-chloro-8-hydroxy-3,4-dihydro-3-R-methylisocoumarin) is the most important mechanism of OTA biodegradation (Figure 2; Loi et al., 2017). Degradation products OTα and L-β-phenylalanine are formed by the hydrolysis of the amide bond via hydrolytic enzymes, such as carboxypeptidase A, carboxypeptidase PJ_1540, protease A, lipase A, ochratoxinase, etc. (Stander et al., 2000; Abrunhosa et al., 2006; Dobritzsch et al., 2014; Liuzzi et al., 2016).
Rodrigues et al. (2009) reported that OTα was non-toxic, or at least 500 times less toxic than OTA. The research of Ferenczi et al. (2014) revealed that the administration of OTA (1 or 10 mg/kg BW for 72 h or 0.5 mg/kg BW for 21 d) to male CD1 mice via oral gavage led to a significant increase of OTA concentration in the blood, transcriptional alterations in OTA-dependent genes (gadd45, gadd153, sulphotransferase, annexinA2, and clusterin) and histopathological changes in the renal cortex. These OTA-induced alterations in male CD1 mice were not observed in the group which OTA was degraded into OTα by treating OTA with Cupriavidus basilensis OR16 for 5 days (Ferenczi et al., 2014). Rodriguez et al. (2011) verified that 0.04 μg/mL of OTA was converted completely to OTα by food-borne Brevibacterium spp. strains (B. casei DSM 20657, DSM 9657, DSM 20658, RM101; B. linens DSM 20425; B. iodinum DSM20626; B. epidermidis DSM 20660) within 10 d. The tested strain, B. casei RM101, could completely cleave OTA even at 40 μg/mL, which is 1,000 times greater than the OTA concentration commonly found in foodstuffs (Rodriguez et al., 2011). Bejaoui et al. (2006) reported that OTα was further degraded into unknown products. Aspergillus niger GX312, A. japonicus AX35, and A. carbonarius SA332 (a weak OTA producer) could convert OTA to OTα by 99, 89, and 83% within 5 days, respectively. Then OTα was converted to an unknown compound after 9 days incubation (Bejaoui et al., 2006). A small amount of authors reported that OTA could be directly degraded into unknown products (Patharajan et al., 2011; Shi et al., 2013, 2014).
Effects of Different Culture Conditions on OTA Biodegradation
The degradation of OTA by the same microorganism in different culture media has different manifestations (Varga et al., 2000, 2005; Abrunhosa et al., 2014). It is worth mentioning that an atoxigenic A. niger CBS 120.49 could completely degrade OTA (2.5 μg/mL) to OTα in solid medium within 5 d, faster than in liquid medium within 7 days (Varga et al., 2000). The degradation product, OTα, was further decomposed to an unknown compound within 7 days in solid media (Varga et al., 2000). Among 55 isolated Rhizopus and Mucor strains, many Rhizopus strains were able to degrade OTA (7.5 μg/mL) to a detection limit below concentration in liquid medium within 10 days (Varga et al., 2005). A. niger CBS 120.49 was able to degrade more than 90% of OTA within 6 d, while R. stolonifer var. stolonifer TJM 8A8 needed 12 days to degrade about 90% of OTA in liquid medium. Only R. stolonifer var. stolonifer TJM 8A8 was able to degrade 96.5% of OTA (7.5 μg/g) in moistened wheat during 10 days incubation (Varga et al., 2005). Pediococcus parvulus UTAD 473 was able to degrade 90% of OTA (1 μg/mL) within 25 h in MRS broth medium, while only degrading 80% of OTA (7 μg/L) after 6 days incubation in grape must. Furthermore, no obvious degradation of OTA (7 μg/L) was observed in synthetic wine (Abrunhosa et al., 2014).
Oxygen is also one of the important factors that affect the growth and reproduction of microorganisms. In addition to certain aerobic microorganisms, some anaerobic microorganisms were also able to degrade OTA (Schatzmayr et al., 2006; Upadhaya et al., 2012). Anaerobic Eubacterium biforme MM11, isolated from swine gut, was able to degrade 77.1% of OTA (0.1 μg/mL) in modified M 98-5 liquid medium within 12 h at 39°C. This strain could completely degrade 1 μg/mL of OTA in solid corn substrate within 24 h at 39°C, which suggests that anaerobic microorganisms might be suitable for the development of feed additives that will function in the targeted animal intestines. It is also worth noting that 26% of the OTA (1 μg/mL) was removed in the negative control (corn) within 24 h at 39°C (Upadhaya et al., 2012). Moreover, anaerobic E. callanderi Due4_11 was able to degrade 95% of OTA (0.2 μg/mL) to OTα within 6 h (Schatzmayr et al., 2006).
Dually Functional Strains of OTA Biodegradation
Some microorganisms were not only able to degrade OTA, but also able to inhibit the biosynthesis of OTA. El Khoury et al. (2017) reported that several actinobacterial strains (Streptomyces AT10, AT8, SN7, MS1, ML5, G10, PT1) were able to degrade 22.83–52.68% of OTA (0.095 μg/mL) within 5 d. At the same time, Streptomyces AT10, AT8, SN7, G10, and PT1 could adsorb 16.07%-33.93% of OTA (0.045 μg/mL) within 1 h. Furthermore, the expression of biosynthesis genes (acOTAnrps, acpks, and acOTApks) and regulatory gene (veA) of OTA in A. carbonarius S402 was inhibited by Streptomyces MS1, ML5, and G10, with the down regulation of 21.0, 11.9, and 11.9% for acOTAnrps, 37.1, 39.0, and 9.0% for acpks, 23.9, 23.0, and 18.3% for acOTApks, and 11.4, 0.0, and 7.0% for veA, respectively (El Khoury et al., 2017).
Some microorganisms have dual functions of OTA degradation and adsorption (Péteri et al., 2007; Shi et al., 2014). Shi et al. (2014) reported that the cell-free supernatant of Bacillus subtilis CW 14 could degrade 97.6% of OTA (6 μg/mL) within 24 h, but no degradation product was detected. Furthermore, 66.6 and 87.9% of OTA (6 μg/mL) was adsorbed by viable and dead B. subtilis CW 14 cells, respectively (Shi et al., 2014). Péteri et al. (2007) verified that Phaffia rhodozyma CBS 5905 was able to degrade 90% of OTA (7.5 μg/mL) within 15 d and adsorb 23% of OTA (3 μg/mL) within 2 h, respectively.
Biodetoxification of OTA by Adsorption
Microorganisms, including actinobacteria, bacteria, filamentous fungi, and yeast, that were able to adsorb OTA are summarized in Table S2. The most important influence factor of OTA adsorption capacity of microorganisms is cell wall components. However, there are controversies among different scholars on specific cell wall components, such as glucogalactans and β-glucans (Ben Taheur et al., 2017), mannoproteins (Caridi et al., 2012), β-glucans and mannans (Pereyra et al., 2015).
Effects of Different Culture Conditions on OTA Adsorption
The adsorption of OTA by the same microorganism in different culture conditions has different manifestations (Armando et al., 2012; Piotrowska et al., 2013; Ben Taheur et al., 2017). Piotrowska et al. (2013) certified that Saccharomyces cerevisiae Syrena LOCK 0201 and S. cerevisiae Malaga LOCK 0173 removed 85.1 and 82.8% of OTA (1 μg/mL) in white grape juice as well as 65.2 and 10.7% of OTA (1 μg/mL) in blackcurrant juice after 10 days incubation, respectively. Armando et al. (2012) simulated mammalian gastrointestinal conditions, Yeast Peptone Dextrose (YPD) broth (pH 2 and pH 7) and YPD with 0.5% bile salts (pH 7), to study the effect of S. cerevisiae RC008, RC009, RC012, and RC016 on OTA (100 μg/mL) adsorption. Results proved that the OTA binding level of S. cerevisiae RC008 and RC009 was 82.3 and 80.2% in YPD broth (pH 2), 74.4 and 78.7% in YPD with 0.5% bile salts (pH 7), 56.7 and 67.1% in YPD broth (pH 7), respectively. The OTA binding ability of S. cerevisiae RC008 and RC009 was significantly stronger in simulated mammalian gastrointestinal conditions than that in YPD broth (pH 7), while the OTA binding ability of S. cerevisiae RC012 and RC016 was not significantly different between simulated mammalian gastrointestinal conditions and YPD broth (pH 7). Mammalian gastrointestinal conditions enhanced the adsorption of S. cerevisiae to OTA, or at least did not reduce OTA-S. cerevisiae interactions (Armando et al., 2012). Ben Taheur et al. (2017) reported that Acetobacter syzygii KFGM1 and Lactobacillus kefiri KFLM3 were able to bind 15 and 15% of OTA in Man Rogosa Sharpe (MRS) medium while 50 and 81% of OTA in milk during 1 d incubation, respectively. Kazachstania servazzii KFGY7 could adsorb 6 and 62% of OTA in yeast extract peptone dextrose (YPD) and in milk during 1 d incubation, respectively. The reason might be that the nutrient components of milk were better than those of MRS or YPD, which facilitated the growth of microorganisms and the biosynthesis of cell wall ingredients concerning OTA adsorption. The OTA adsorption mechanism of L. kefiri KFLM3 and K. servazzii KFGY7 was possibly involved in the function of hydrosoluble glucogalactan exopolysaccharides and β-(1,3 and 1,6)-D-glucans of the cell wall, respectively (Ben Taheur et al., 2017).
Effects of Different Statuses on OTA Adsorption
The adsorption of OTA by the same microorganism but in different statuses (viable/dead) has similar or different manifestations (Bejaoui et al., 2004, 2005; Péteri et al., 2007; Mateo et al., 2010; Fiori et al., 2014; Piotrowska, 2014). Péteri et al. (2007) reported that viable and dead Phaffia rhodozyma CBS 5905 were able to remove 23 and 45% of OTA (3 μg/mL) within 2 h, respectively. Mateo et al. (2010) demonstrated that the OTA adsorption capacity of Oenococcus oeni 6G and O. oeni 124 M was not significantly different between viable and dead cells after 30 min incubation with 2 μg/L of OTA. Fiori et al. (2014) tested the OTA adsorption capacity of viable/dead yeast strains. Results demonstrated that viable and dead Candida friedrichii 778, Candida intermedia 235, Lachancea thermotolerans 751, Cyberlindnera jadinii 273 could adsorb 70, 73, 75, and 0% of OTA (0.02 μg/mL) and 72, 74, 84, and 82% of OTA within 8 days, respectively. The OTA adsorption capacity varied greatly between viable and dead yeast strain C. jadinii 273 (Fiori et al., 2014). Bejaoui et al. (2005) reported that the OTA adsorption ability of dead A. niger GX312, A. carbonarius SA332 (a weak OTA producer), and A. Japonicus AX35, with the OTA (2 μg/mL) reduction of 47.5, 66.5, and 41.5% within 2 h, was at least 10% higher than that of viable A. niger GX312, A. carbonarius SA332 (a weak OTA producer), and A. Japonicus AX35, with the OTA (2 μg/mL) reduction of 30, 55, and 30% within 2 h. While viable or dead A. niger GX312, A. carbonarius SA332 (a weak OTA producer), and A. Japonicus AX35 displayed the same adsorption ability to low concentration of OTA that 80% of OTA (0.01 μg/mL) was removed within 2 h (Bejaoui et al., 2005). Bejaoui et al. (2004) demonstrated that viable and dead S. cerevisiae LALVIN Rhône 2056 was able to remove 17 and 75% of OTA (2 μg/mL) within 2 h in liquid yeast peptone glucose (YPG) medium, respectively. Furthermore, the OTA adsorption ability was not significantly influenced by different treatment methods that lead to microbial cell death, since no significant difference was observed between heat and acid treatments of S. cerevisiae LALVIN Rhône 2056 cells. Heat treatment resulted in the formation of Maillard reaction products, the denaturation of proteins, the reduction of cell wall thickness of peptidoglycan, or the enlargement of pore size, which may be responsible for more exposure of OTA adsorption sites than viable cells. Acid treatment led to the formation of monomers and aldehydes products via breaking down polysaccharides, the reduction of cell wall thickness of peptidoglycan, or the enlargement of pore size, which may be responsible for more exposure of OTA adsorption sites as well (Bejaoui et al., 2004). Piotrowska (2014) verified that the dead Lactobacillus plantarum LOCK 0862, L. brevis LOCK 0845, and L. sanfranciscensis LOCK 0866 could reduce 46.29–59.82% of OTA (1 μg/mL) within 30 min in PBS buffer, but the alive L. plantarum LOCK 0862, L. brevis LOCK 0845, and L. sanfranciscensis LOCK 0866 needed 24 h to remove 14.80–26.42% of OTA (1 μg/mL) in PBS buffer. The adsorption of OTA to the surface structures of the lactic acid bacteria cell wall is influenced by hydrophobic, acceptor/acidic, and donor/basic properties of the cell surface (Piotrowska, 2014).
Effects of Microbial Cell Wall Components on OTA Adsorption
In addition, some researchers have specifically studied the OTA adsorption ability of microbial cell walls (Pereyra et al., 2015). Two commercial yeast cell walls (YCW) with different β-glucans/mannans percentages, YCW1 (23.3% of polysaccharides containing 17.4% of β-glucans and 5.9% of mannans) and YCW2 (44.0% of polysaccharides containing 23.0% of β-glucans and 21.0% of mannans), were used to evaluate their OTA adsorption ability in simulated stomachal conditions of monogastric animals by Pereyra et al. (2015). Results proved that the OTA adsorption ability was not significantly influenced by different percentages of polysaccharides or β-glucans/mannans (Pereyra et al., 2015).
Effects of Strains and Their Statuses on the Firmness of OTA Adsorption
The firmness of OTA adsorption was strain-specific (Caridi et al., 2012; Petruzzi et al., 2015). Petruzzi et al. (2015) reported that S. cerevisiae W28 and W46 was able to bind 34.51–70.17% and 42.79–76.44% of OTA (2 μg/L) in grape must, respectively. But, the binding of S. cerevisiae-OTA was reversible with 80%-85% of the initially binding OTA releasing back into the washing solution. The binding ability of S. cerevisiae-OTA complex varied among different kinds of strains. The S. cerevisiae W13 could remove 30.69–53.79% of OTA (2 μg/L) in grape must, but its releasing-back percentage was 55%, which was significantly lower than that of S. cerevisiae W28 and W46 (Petruzzi et al., 2015). Caridi et al. (2012) verified that there is a great difference between descendants and their parents (S. cerevisiae TP5 and TT173) in the homogeneity of OTA adsorption capacity. The different content of mannoproteins might contribute to the difference of OTA adsorption capacity of yeast cells, because OTA and mannoproteins were linked through ionic and electrostatic interactions (Caridi et al., 2012).
The firmness of OTA adsorption by the same microorganism in different statuses (viable/dead) was also different (Piotrowska, 2012). The bond between microbial cells and OTA was partially reversible. Piotrowska (2012) reported that the complex of viable cells-OTA was firmer than the complex of dead cells-OTA, with 11 and 22% of initially binding OTA releasing back into PBS buffer, respectively. Protoplasts of S. cerevisiae BS lost the capacity to bind OTA, indicating that the components of the cell wall were strongly associated with the adsorption of OTA (Piotrowska, 2012).
Biodetoxification of OTA by Enzymes
Enzymes, including crude and purified, that were able to cleave OTA, are summarized in Table S3.
The first report about the biodetoxification of OTA took place in 1969 (Pitout, 1969), which was only four years after the discovery of OTA (Van Der Merwe et al., 1965). It used bovine pancreatic carboxypetidase A (CPA) to cleave OTA to OTα (Pitout, 1969).
Abrunhosa et al. (2006) reported that Ancex, a crude enzyme isolated from A. niger MUM 03.58, was able to degrade 99.8% of OTA (1 μg/mL) to OTα after 25 h incubation at pH 7.5 and 37°C. Commercially purified enzymes did not degrade nearly as much. For example, 87.3 and 43.4% of OTA was degraded by Protease A and Pancreatin after 25 h incubation at pH 7.5 and 37°C, respectively (Abrunhosa et al., 2006). Furthermore, another commercially purified enzyme, Prolyve PAC, was able to degrade only 3% of OTA (1 μg/mL) to OTα after 25 h incubation at pH 3 (optimal pH) and 37°C (Abrunhosa et al., 2006). A crude metalloenzyme, with 12.8 times higher OTA hydrolytic activity than that of CPA at pH 7.5 and 37°C, was isolated from A. niger by Abrunhosa and Venâncio (2007). Finally, an OTA hydrolytic activity of 36 U/mg was obtained from the purified enzyme (Abrunhosa and Venâncio, 2007). Cho et al. (2016) reported that 97.5 and 91.3% of OTA (0.04 μg/mL) was removed by crude enzymes of A. tubingensis M036 and M074 at pH 5 and 25°C within 24 h, respectively.
Dobritzsch et al. (2014) reported that the purified recombinant ochratoxinase was about 600 times more efficiently hydrolyzed OTA than CPA at an optimal pH of CPA (pH 7.5). The optimal OTA-degrading activity of this novel ochratoxinase was obtained at 66°C and about pH 6 (Dobritzsch et al., 2014).
In general, although researchers have isolated many OTA biodegradable microorganisms, there is still a lack of isolation and purification of enzymes and performance studies among them. This may be mainly due to the relatively small number of interdisciplinary studies, and relatively few experts familiar with the relevant studies of both microorganisms and enzymes, thus limiting the subsequent in-depth study of abundant microorganisms with excellent biodegradability.
Industrial Application Prospects of OTA Biodetoxification
It has been reported that a large number of patented microorganisms have great application prospects in food and feed industries (Guan et al., 2009; Jiang et al., 2016a,b,c,d). After 72 h incubation with Bacillus licheniformis MZH-11, 0.1, 0.5, and 5 μg/g of OTA in corn flour was degraded by 84.4,78.3, and 73.5%, respectively (Guan et al., 2009). Although the concentration of OTA in corn flour increased by 10-fold from 0.5 to 5 μg/g, the decreased degradation rate of OTA from 78.3 to 73.5% was very little, which indicated the great application prospects of B. licheniformis MZH-11 in the feed industry (Guan et al., 2009). Jiang et al. (2016b) reported that 0.02 μg/g of OTA in corn-soybean feed was degraded by 71% after 72 h incubation with Stenotrophomonas sp. CW117. Moreover, 0.02 μg/g of OTA in corn-soybean feed was degraded by 48.3, 53.2, and 68.7% following 48 h incubation with Luteimonas sp. CW574 (Jiang et al., 2016d), Silanimonas sp. CW282 (Jiang et al., 2016c), and Lysobacter sp. CW239 (Jiang et al., 2016a), respectively.
It is essential for industrial application of strains to maintain good performance during processing, such as lyophilization. Schatzmayr et al. (2006) used an in vitro model with whole pieces of pig gut to evaluate the OTA-degradation activity of lyophilized powders of Trichosporon mycotoxinivorans (MTV, 115) and Stenotrophomonas nitritreducens 041-9 strains. Results revealed that the lyophilized powders of T. mycotoxinivorans (MTV, 115) or S. nitritreducens 041-9 could degrade about 90% of OTA (0.4 μg/mL) within 6 h, which displayed great potential to be applied as OTA-detoxifying feed additive (Schatzmayr et al., 2006). Due to the excellent OTA-detoxification performance of T. mycotoxinivorans (MTV, 115), it was made into a product named Mycofix® PlusMTVINSIDE by Biomin GmbH (Austria) (Hanif et al., 2008). Hanif et al. (2008) added 1‰ and 2‰ Mycofix® PlusMTVINSIDE (containing 6. 0 × 108 count T. mycotoxinivorans MTV cells/g) to the feed containing OTA (0.5 and 1 μg/g) for studying the attenuating effects of Mycofix® PlusMTVINSIDE on the undesirable effects of OTA in 1-day-old broiler chicks over a 42-days period. Results confirmed that OTA induced adverse effects, such as poor feed conversion ratio, depressed body weight gain, increased levels of serum enzymes (lactate dehydrogenase, aspartate aminotransferase, and γ-glutamyltranspeptidase) and significant histopathological changes in bursa of fabricius, spleen, liver, and kidney, were appreciably attenuated by Mycofix® PlusMTVINSIDE (Hanif et al., 2008). Although Mycofix® PlusMTVINSIDE has many advantages in OTA detoxification, T. mycotoxinivorans was recognized in 2009 as a possible novel human pathogen that is related to cystic fibrosis (Hickey et al., 2009), suggesting that the secondary metabolites of T. mycotoxinivorans are extremely complicated. Therefore, T. mycotoxinivorans deserves further investigations to reduce possible deleterious effects and should be used with caution in food or feed industries. It is noteworthy that the application of microorganisms to the food and feed industries must be cautious, with particular attention to risks for human health (Spano and Capozzi, 2011; Capozzi et al., 2017).
It has also been reported that many microorganisms with good OTA adsorption ability have great application prospects in food and feed industries (Moruno et al., 2005; Csutorás et al., 2013; Farbo et al., 2016). Csutorás et al. (2013) used macro-scale experiments (16 L glass balloons in wine cellar) to simulate a real wine industrial system. During a 90-day fermentation process, OTA (4 μg/mL) was adsorbed to S. cerevisiae by 73, 85, and 90% in white, rose, and red wine musts, respectively (Csutorás et al., 2013). Moruno et al. (2005) used S. cerevisiae 169-involving fermentation lees of red and white grape musts to treat OTA-containing red wine samples. Results certified that 4.12 and 7.09 μg/L of OTA was adsorbed by 58.7 and 71.4% in 80 d red wine fermentation with red lees and white lees, respectively (Moruno et al., 2005). Farbo et al. (2016) immobilized Candida intermedia 253 yeast cells into magnetic calcium alginate beads to adsorb OTA in commercial grape juice. Results showed that more than 80% of OTA (0.02 μg/g) was adsorbed within 48 h of incubation, while OTA was slowly released back into the commercial grape juice from calcium alginate beads in the following phases (72–120 h). Thus, the authors developed a glass chromatography column-containing prototype bioreactor encapsulating C. intermedia 253 yeast cells in calcium alginate beads to decontaminate OTA in liquid matrices. The concentration of OTA (0.02 μg/g) in commercial grape juice was reduced by 21 and 57% via the first column filtration step and the fourth column filtration step, respectively (Farbo et al., 2016). Furthermore, three industrial yeast by-products including EX16 (a vinasse containing 16% liquid yeast cell walls), LEC (a dry yeast cell wall fraction), and BETA (a dried purified β-glucans of yeast cell wall fraction) were used to adsorb OTA by Ringot et al. (2007). Both non-polar and polar non-covalent interactions were involved in the plentiful OTA adsorption onto LEC. The non-polar interactions concerned the interactions between hydrophobic amino acids of LEC and aromatic rings of OTA. The polar interactions could be interpreted by four different complementary patterns as follows: (a) electrostatic π-π interactions between aromatic amino acids of LEC and aromatic rings of OTA, (b) electrostatic ionic interactions between basic amino acids of LEC and carboxylic group of OTA, (c) hydrogen bond interactions between donor groups of LEC and receptor aromatic rings of OTA, (d) hydrogen bond interactions between donor groups (phenol and amide group of OTA) and receptor groups of LEC. The relative importance of these interactions between OTA and LEC still needs to be further explored (Ringot et al., 2005).
Furthermore, it has been reported that many microorganisms could detoxify OTA, but the adsorption and/or biodegradation mechanisms of OTA elimination were not reported (not shown in Tables S1–S3). Kapetanakou et al. (2012) applied strains mixtures (107 cfu/mL) with 16 yeasts isolated from different batches of wine and 29 bacteria isolated from fermented flour, basil, sourdough, sausage, and aniseed to degrade OTA (0.1 μg/mL) in grape juice, red wine, and beer at 25°C for 5 days. Results shown OTA reduction was 32, 22, and 12% in grape juice, red wine, and beer, respectively (Kapetanakou et al., 2012). The licensed probiotic preparation, including L. paracasei LOCK 0920, L. plantarum LOCK 0945, L. brevis LOCK 0944, S. cerevisiae LOCK 0140, and Yucca schidigera extract, was applied to reduce OTA in broiler feed by Slizewska and Piotrowska (2014). The 1 and 5 mg/kg of OTA was reduced by 73 and 55% in feed after 6 h fermentation with the probiotic preparation, respectively. At the same time, aerobic spore-bearing bacteria were inhibited by the probiotic preparation (Slizewska and Piotrowska, 2014). Cai et al. (2014) patented a yeast strain isolated from pig feces and nearby soil, Kluyveromyces marxianus C2, which could reduce 82.3% of OTA (0.5 μg/mL) in YPD medium and 83.7% of OTA (0.082 μg/g) in moldy corn feed, respectively.
In addition, it has been reported that a number of enzymes originating from some microorganisms with good OTA degradation ability have great application prospects in food and feed industries (Abrunhosa et al., 2006; Yu et al., 2015). Amidase 2, a patented food or feed additive, was cloned from A. niger to degrade OTA to OTα by Yu et al. (2015). After 2.5 h incubation with amidase 2 at 40°C, the content of OTA was reduced from 47 ng/mL to less than 2 ng/mL in milk (Yu et al., 2015). After 20 h incubation with amidase 2, 38 μg/mL of OTA was reduced to less than 2 ng/mL in corn flour and to 6.6 μg/mL in corn soy based feed, respectively (Yu et al., 2015). Because of the low toxigenicity, A. niger is known as an GRAS (generally regarded as safe) strain, which, to a certain degree, indicates that A. niger-derivatived enzymes have great safety (Varga et al., 2000). Abrunhosa et al. (2006) reported that Ancex, a crude enzyme isolated from A. niger MUM 03.58, was able to degrade 87.9% of OTA (1 μg/mL) to OTα after 3 h incubation at pH 7.5 and 50°C.
Future Directions
Since the discovery of which carboxypeptidase A could degrade OTA in 1969, studies on OTA biodetoxification have been carried out for nearly 50 years. However, there are still many problems that remain to be solved. For example, compared to a single-OTA exposure environment, is the biodetoxification of strains equally effective in a multimycotoxins environment? Moreover, is the strain equally effective in a variety of food or feed complex systems? Furthermore, is it possible to screen a large number of strains in pre-harvest period to prevent OTA biosynthesis and in post-harvest period to biodetoxify OTA at the same time? In addition, at present, the vast majority of studies have been confined to the traditional isolation and screen of strains from different sources. It is necessary to make better use of transgenic technology in mutant strains to obtain better biodetoxification performance of OTA or OTA combined with several different mycotoxins. As the prevailing situation is that a myriad of mycotoxins coexist in the food or/and feed systems, obtaining high-performance strains that simultaneously biodegrade or/and adsorb several mycotoxins is bound to be a trend in the future. Of course, achieving these must be based on the clear studies concerning the biodetoxification mechanisms and genes of several different mycotoxins.
Conclusion
On the basis of the development of about 50 years, researchers have identified a good deal of microorganisms, including actinobacteria, bacteria, filamentous fungi, and yeast, that could degrade and/or adsorb OTA. The degradation of OTA to non-toxic or less toxic OTα is the most important OTA biodegradation mechanism. The OTA adsorption capacity of microorganisms is possibly influenced by the glucogalactan exopolysaccharide, β-(1,3 and 1,6)-D-glucans, mannoproteins, and mannans of the cell wall and the hydrophobic, acceptor/acidic, and donor/basic properties of the cell surface via ionic and electrostatic interactions. The formation of Maillard reaction products, monomers and aldehydes products, the denaturation of proteins, the reduction of cell wall thickness of peptidoglycan, or the enlargement of pore size of dead cells (heat or acid treatment) resulted in more exposure of OTA adsorption sites than that of viable cells, which might be responsible for higher OTA adsorption. Numerous OTA degradation enzymes were isolated or cloned from bacteria, filamentous fungi, yeast, and animal pancreas. A large number of microorganisms with good OTA degradation and/or adsorption ability, as well as some OTA degradation enzymes, have great application prospects in food and feed industries. It is noteworthy that the application of microorganisms to the food and feed industries must be cautious, with particular attention to their safety.
Author Contributions
WC and CL wrote the manuscript. JY, ZZ, and XL searched the databases. BZ offered suggestions and polished language. YS, XhL, YW, and YL offered suggestions. XS modified the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the District Program of National Natural Science Foundation of China (31460426), the Youth Program of National Natural Science Foundation of China (31601577), the Excellent Youth Talents of Zunyi Medical University (17zy-006), the cooperation projects of the Science and Technology Department of Guizhou Province (LH[2014]7546), the project of Zunyi Science and Technology Bureau (201718), the project of Zunyi Medical University (ZMKD2013-022), the project of Beijing Vocational College of Agriculture (XY-YT-16-44), and the supporting discipline construction fund of Zunyi Medical University Ph.D. authorization. The authors also appreciate Mr. Paul Snyder, Ms. Jennie Padlo, and Dr. Jifen Li of University of Pennsylvania for language polishing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01386/full#supplementary-material
References
Abrunhosa, L., Inês, A., Rodrigues, A. I., Guimarães, A., Pereira, V. L., Parpot, P., et al. (2014). Biodegradation of Ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 188, 45–52. doi: 10.1016/j.ijfoodmicro.2014.07.019
Abrunhosa, L., Paterson, R. R., and Venâncio, A. (2010). Biodegradation of Ochratoxin A for food and feed decontamination. Toxins (Basel) 2, 1078–1099. doi: 10.3390/toxins2051078
Abrunhosa, L., Santos, L., and Venâncio, A. (2006). Degradation of Ochratoxin A by proteases and by a crude enzyme of Aspergillus niger. Food Biotechnol. 20, 231–242. doi: 10.1080/08905430600904369
Abrunhosa, L., Serra, R., and Venâncio, A. (2002). Biodegradation of Ochratoxin A by fungi isolated from grapes. J. Agric. Food Chem. 50, 7493–7496. doi: 10.1021/jf025747i
Abrunhosa, L., and Venâncio, A. (2007). Isolation and purification of an enzyme hydrolyzing Ochratoxin A from Aspergillus niger. Biotechnol. Lett. 29, 1909–1914. doi: 10.1007/s10529-007-9479-2
Abrunhosa, L., Venâncio, A., and Teixeira, J. A. (2011). Optimization of process parameters for the production of an OTA-hydrolyzing enzyme from Aspergillus niger under solid-state fermentation. J. Biosci. Bioeng. 112, 351–355. doi: 10.1016/j.jbiosc.2011.06.016
Angioni, A., Caboni, P., Garau, A., Farris, A., Orro, D., Budroni, M., et al. (2007). In vitro interaction between Ochratoxin A and different strains of Saccharomyces cerevisiae and Kloeckera apiculata. J. Agric. Food Chem. 55, 2043–2048. doi: 10.1021/jf062768u
Armando, M. R., Pizzolitto, R. P., Dogi, C. A., Cristofolini, A., Merkis, C., Poloni, V., et al. (2012). Adsorption of Ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness. J. Appl. Microbiol. 113, 256–264. doi: 10.1111/j.1365-2672.2012.05331.x
Bejaoui, H., Mathieu, F., Taillandier, P., and Lebrihi, A. (2004). Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains. J. Appl. Microbiol. 97, 1038–1044. doi: 10.1111/j.1365-2672.2004.02385.x
Bejaoui, H., Mathieu, F., Taillandier, P., and Lebrihi, A. (2005). Conidia of black Aspergilli as new biological adsorbents for Ochratoxin A in grape juices and musts. J. Agric. Food Chem. 53, 8224–8229. doi: 10.1021/jf051029v
Bejaoui, H., Mathieu, F., Taillandier, P., and Lebrihi, A. (2006). Biodegradation of Ochratoxin A by Aspergillus section Nigri species isolated from French grapes: a potential means of Ochratoxin A decontamination in grape juices and musts. FEMS Microbiol. Lett. 255, 203–208. doi: 10.1111/j.1574-6968.2005.00073.x
Bhat, P. V., Pandareesh Khanum, F., and Tamatam, A. (2016). Cytotoxic effects of Ochratoxin A in Neuro-2a cells: role of oxidative stress evidenced by N-acetylcysteine. Front. Microbiol. 7:1142. doi: 10.3389/fmicb.2016.01142
Bizaj, E., Mavri, J., Cuš, F., and Raspor, A. (2016). Removal of Ochratoxin A in Saccharomyces cerevisiae Liquid Cultures. S. Afr. J. Enol. Vitic. 30, 151–155. doi: 10.21548/30-2-1436
Böhm, J., Grajewski, J., Asperger, H., Cecon, B., Rabus, B., and Razzazi, E. (2000). Study on biodegradation of some A- and B-trichothecenes and Ochratoxin A by use of probiotic microorganisms. Mycotoxin Res. 16(Suppl. 1), 70–74. doi: 10.1007/bf02942985
Bondy, G. S., Caldwell, D. S., Aziz, S. A., Coady, L. C., Armstrong, C. L., Curran, I. H., et al. (2015). Effects of chronic Ochratoxin A exposure on p53 heterozygous and p53 homozygous mice. Toxicol. Pathol. 43, 715–729. doi: 10.1177/0192623314568391
Bui-Klimke, T. R., and Wu, F. (2015). Ochratoxin A and human health risk: a review of the evidence. Crit. Rev. Food Sci. Nutr. 55, 1860–1869. doi: 10.1080/10408398.2012.724480
Cai, H., Chen, H., Chen, Z., Han, J., Jin, M., Li, F., et al. (2014). New Marx Kluyveromyces Yeast Comprising Kluyveromyces Marxianus C2, Useful for Preparing Animal Feed Additive and Acute Poisoning Antidotes in Fungi, and Degradation of Aflatoxin B1, Corn Zearalenone and Ochratoxin A. Beijing: State Intellectual Property Office.
Capozzi, V., Fragasso, M., Romaniello, R., Berbegal, C., Russo, P., and Spano, G. (2017). Spontaneous food fermentations and potential risks for human health. Fermentation 3:49. doi: 10.3390/fermentation3040049
Cappozzo, J., Jackson, L., Lee, H. J., Zhou, W., Al-Taher, F., Zweigenbaum, J., et al. (2017). Occurrence of Ochratoxin A in infant foods in the United States. J. Food Prot. 80, 251–256. doi: 10.4315/0362-028x.jfp-16-339
Caridi, A., Galvano, F., Tafur, A., and Ritieni, A. (2006). In-vitro screening of Saccharomyces strains for Ochratoxin A removal from liquid medium. Ann. Microbiol. 56, 135–137. doi: 10.1007/BF03174994
Caridi, A., Sidari, R., Pulvirenti, A., Meca, G., and Ritieni, A. (2012). Ochratoxin A adsorption phenotype: an inheritable yeast trait. J. Gen. Appl. Microbiol. 58, 225–233. doi: 10.2323/jgam.58.225
Cecchini, F., Morassut, M., Garcia Moruno, E., and Di Stefano, R. (2006). Influence of yeast strain on Ochratoxin A content during fermentation of white and red must. Food Microbiol. 23, 411–417. doi: 10.1016/j.fm.2005.08.003
Chang, X., Wu, Z., Wu, S., Dai, Y., and Sun, C. (2015). Degradation of Ochratoxin A by Bacillus amyloliquefaciens ASAG1. Food Addit. Contam. A 32, 564–571. doi: 10.1080/19440049.2014.991948
Cho, S. M., Jeong, S. E., Lee, K. R., Sudhani, H. P., Kim, M., Hong, S. Y., et al. (2016). Biodegradation of Ochratoxin A by Aspergillus tubingensis Isolated from Meju. J. Microbiol. Biotechnol. 26, 1687–1695. doi: 10.4014/jmb.1606.06016
Cserháti, M., Kriszt, B., Krifaton, C., Szoboszlay, S., Háhn, J., Tóth, S., et al. (2013). Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 166, 176–185. doi: 10.1016/j.ijfoodmicro.2013.06.002
Csutorás, C., Rácz, L., Rácz, K., Futo, P., Forgó, P., and Kiss, A. (2013). Monitoring of Ochratoxin A during the fermentation of different wines by applying high toxin concentrations. Microchem. J. 107, 182–184. doi: 10.1016/j.microc.2012.07.001
Dai, Q., Zhao, J., Qi, X., Xu, W., He, X., Guo, M., et al. (2014). MicroRNA profiling of rats with Ochratoxin A nephrotoxicity. BMC Genomics 15, 333. doi: 10.1186/1471-2164-15-333
De Bellis, P., Tristezza, M., Haidukowski, M., Fanelli, F., Sisto, A., Mulè, G., et al. (2015). Biodegradation of Ochratoxin A by bacterial strains isolated from vineyard soils. Toxins (Basel) 7, 5079–5093. doi: 10.3390/toxins7124864
Deberghes, P., Betbeder, A. M., Boisard, F., Blanc, R., Delaby, J. F., Krivobok, S., et al. (1995). Detoxification of Ochratoxin A, a food contaminant: prevention of growth of Aspergillus ochraceus and its production of Ochratoxin A. Mycotoxin Res. 11, 37–47. doi: 10.1007/bf03192060
de Felice, D. V., Solfrizzo, M., De Curtis, F., Lima, G., Visconti, A., and Castoria, R. (2008). Strains of Aureobasidium pullulans can lower Ochratoxin A contamination in wine grapes. Phytopathology 98, 1261–1270. doi: 10.1094/phyto-98-12-1261
Del Prete, V., Rodriguez, H., Carrascosa, A. V., de las Rivas, B., Garcia-Moruno, E., and Muñoz, R. (2007). In vitro removal of Ochratoxin A by wine lactic acid bacteria. J. Food Prot. 70, 2155–2160.
Dobritzsch, D., Wang, H., Schneider, G., and Yu, S. (2014). Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 462, 441–452. doi: 10.1042/bj20140382
Duarte, S. C., Pena, A., and Lino, C. M. (2010). A review on Ochratoxin A occurrence and effects of processing of cereal and cereal derived food products. Food Microbiol. 27, 187–198. doi: 10.1016/j.fm.2009.11.016
el Khoury, A., and Atoui, A. (2010). Ochratoxin A: general overview and actual molecular status. Toxins (Basel) 2:461. doi: 10.3390/toxins2040461
Engelhardt, G. (2002). Degradation of Ochratoxin A and B by the white rot fungus pleurotus ostreatus. Mycotoxin Res. 18, 37–43. doi: 10.1007/bf02946138
Fanelli, F., Chiara, M., Liuzzi, V. C., Haidukowski, M., Tristezza, M., Caterina, M., et al. (2015). Draft genome sequence of Acinetobacter sp neg1 capable of degrading Ochratoxin A. FEMS Microbiol. Lett. 362:fnv004. doi: 10.1093/femsle/fnv004
Farbo, M. G., Urgeghe, P. P., Fiori, S., Marceddu, S., Jaoua, S., and Migheli, Q. (2016). Adsorption of Ochratoxin A from grape juice by yeast cells immobilised in calcium alginate beads. Int. J. Food Microbiol. 217, 29–34. doi: 10.1016/j.ijfoodmicro.2015.10.012
Ferenczi, S., Cserháti, M., Krifaton, C., Szoboszlay, S., Kukolya, J., Szoke, Z., et al. (2014). A new Ochratoxin A biodegradation strategy using Cupriavidus basilensis Or16 Strain. PLoS ONE 9:e109817. doi: 10.1371/journal.pone.0109817
Fiori, S., Urgeghe, P. P., Hammami, W., Razzu, S., Jaoua, S., and Migheli, Q. (2014). Biocontrol activity of four non- and low-fermenting yeast strains against Aspergillus carbonarius and their ability to remove Ochratoxin A from grape juice. Int. J. Food Microbiol. 189, 45–50. doi: 10.1016/j.ijfoodmicro.2014.07.020
Freire, L., Passamani, F. R. F., Thomas, A. B., Nassur, R. C. M. R., Silva, L. M., Paschoal, F. N., et al. (2017). Influence of physical and chemical characteristics of wine grapes on the incidence of Penicillium and Aspergillus fungi in grapes and Ochratoxin A in wines. Int. J. Food Microbiol. 241, 181–190. doi: 10.1016/j.ijfoodmicro.2016.10.027
Fuchs, S., Sontag, G., Stidl, R., Ehrlich, V., Kundi, M., and Knasmüeller, S. (2008). Detoxification of patulin and Ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 46, 1398–1407. doi: 10.1016/j.fct.2007.10.008
Gallo, A., Ferrara, M., and Perrone, G. (2013). Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins (Basel) 5, 717. doi: 10.3390/toxins5040717
Gallo, A., Ferrara, M., and Perrone, G. (2017). Recent advances on the molecular aspects of Ochratoxin A biosynthesis. Curr. Opin. Food Sci. 17, 49–56. doi: 10.1016/j.cofs.2017.09.011
Gil-Serna, J., Patiño, B., Cortés, L., González-Jaén, M. T., and Vázquez, C. (2011). Mechanisms involved in reduction of Ochratoxin A produced by Aspergillus westerdijkiae using Debaryomyces hansenii CYC 1244. Int. J. Food Microbiol. 151, 113–118. doi: 10.1016/j.ijfoodmicro.2011.08.012
Guan, S., Ji, C., Liang, Z., Li, J., Niu, T., and Tian, J. (2009). Sieving Strain for Synchronously Degrading aflatoxin B1 (AFB1) and Ochratoxin A (OTA), Comprises Sieving Strains Capable of Degrading Coumarin, Sieving Strains Capable of Degrading AFB1 and OTA, and Using Multiple Strains. Beijing: State Intellectual Property Office.
Hanif, N. Q., Muhammad, G., Siddique, M., Khanum, A., Ahmed, T., Gadahai, J. A., et al. (2008). Clinico-pathomorphological, serum biochemical and histological studies in broilers fed Ochratoxin A and a toxin deactivator (Mycofix® Plus). Br. Poult. Sci. 49, 632–642. doi: 10.1080/00071660802295183
Harris, J. P., and Mantle, P. G. (2001). Biosynthesis of ochratoxins by Aspergillus ochraceus. Phytochemistry 58, 709–716. doi: 10.1016/S0031-9422(01)00316-8
Hathout, A. S., and Aly, S. E. (2014). Biological detoxification of mycotoxins: a review. Ann. Microbiol. 64, 905–919. doi: 10.1007/s13213-014-0899-7
Hickey, P. W., Sutton, D. A., Fothergill, A. W., Rinaldi, M. G., Wickes, B. L., Schmidt, H. J., et al. (2009). Trichosporon mycotoxinivorans, a novel respiratory pathogen in patients with cystic fibrosis. J. Clin. Microbiol. 47, 3091–3097. doi: 10.1128/jcm.00460-09
Hong, J. T., Park, K. L., Han, S. Y., Park, K. S., Kim, H. S., Oh, S. D., et al. (2000). Effects of Ochratoxin A on cytotoxicity and cell differentiation in cultured rat embryonic cells. J. Toxicol. Env. Heal. A 61, 609–621. doi: 10.1080/00984100050194126
Huff, W. E., and Hamilton, P. B. (1979). Mycotoxins-their biosynthesis in fungi: ochratoxins-metabolites of combined pathways. J. Food Prot. 42, 815–820. doi: 10.4315/0362-028X-42.10.815
Hwang, C.-A., and Draughon, F. A. (1994). Degradation of Ochratoxin A by Acinetobacter calcoaceticus. J. Food Prot. 57, 410–414. doi: 10.4315/0362-028x-57.5.410
IARC (1993). “IARC monographs on the evaluation of carcinogenic risks to humans, some naturally occurring substances: food items and constituents, heterocyclic aromatic amines and mycotoxins,” in IARC Science Publications (Lyon: International Agency for Research on Cancer), 489–521.
Jackson, L. S., and Ryu, D. (2017). Summary of the ACS symposium on public health perspectives of mycotoxins in Food. J. Agric. Food Chem. 65, 7017–7020. doi: 10.1021/acs.jafc.7b02909
Jiang, N., Wang, X., Zhang, Z., and Zhou, Y. (2016a). New Lysobacter sp. CW239 Used in Biodegradation, and in Mildew Food Raw Material or Feed Detoxification Treatment. Beijing: State Intellectual Property Office.
Jiang, N., Wang, X., Zhang, Z., Zhou, Y., and Wu, Y. (2016b). New Stenotrophomonas Species Useful in Preparing Biodegradable Agents for Detoxification Treatment of Moldy Food Materials and Animal Feed. Beijing: State Intellectual Property Office.
Jiang, N., Wang, X., Zhou, Y., and Wei, C. (2016c). New Silanimonas sp. (CW282) Useful for Preparing Aflatoxin B1 and Ochratoxin A Bifunctional Biodegradable Agent. Beijing: State Intellectual Property Office.
Jiang, N., Wang, X., Zhou, Y., Wei, C., and Wu, Y. (2016d). New Luteimonas Species Useful in Preparing Biodegradable Agents for Detoxification Treatment of Moldy Food Materials and Animal Feed. Beijing: State Intellectual Property Office.
Kabak, B., and Dobson, A. D. (2009). Biological strategies to counteract the effects of mycotoxins. J. Food Prot. 72, 2006–2016. doi: 10.4315/0362-028X-72.9.2006
Kapetanakou, A. E., Kollias, J. N., Drosinos, E. H., and Skandamis, P. N. (2012). Inhibition of A. carbonarius growth and reduction of Ochratoxin A by bacteria and yeast composites of technological importance in culture media and beverages. Int. J. Food Microbiol. 152, 91–99. doi: 10.1016/j.ijfoodmicro.2011.09.010
Khoury, R. E., Mathieu, F., Atoui, A., Kawtharani, H., Khoury, A. E., Afif, C., et al. (2017). Ability of soil isolated actinobacterial strains to prevent, bind and biodegrade Ochratoxin A. Toxins (Basel) 9:222. doi: 10.3390/toxins9070222
Lee, H. J., and Ryu, D. (2017). Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: public health perspectives of their co-occurrence. J. Agric. Food Chem. 65, 7034–7051. doi: 10.1021/acs.jafc.6b04847
Leonardo, P., Antonio, B., Rosaria, C. M., Barbara, S., Vittorio, C., and Milena, S. (2017). A Focus on Quality and Safety Traits of Saccharomyces cerevisiae Isolated from Uva di Troia Grape Variety. J. Food Sci. 82, 124–133. doi: 10.1111/1750-3841.13549
Li, X., Zhao, L., Fan, Y., Jia, Y., Sun, L., Ma, S., et al. (2014). Occurrence of mycotoxins in feed ingredients and complete feeds obtained from the Beijing region of China. J. Anim. Sci. Biotechnol. 5:37. doi: 10.1186/2049-1891-5-37
Liew, W. P., and Mohd-Redzwan, S. (2018). Mycotoxin: its impact on gut health and microbiota. Front. Cell. Infect. Microbiol. 8:60. doi: 10.3389/fcimb.2018.00060
Lim, C. W., Yoshinari, T., Layne, J., and Chan, S. H. (2015). Multi-mycotoxin screening reveals separate occurrence of aflatoxins and Ochratoxin A in Asian rice. J. Agric. Food Chem. 63, 3104–3113. doi: 10.1021/acs.jafc.5b00471
Liu, Y., Wang, Y., Xing, F., and Zhao, Y. (2016). Use of Copper Pseudomonas aeruginosa and/or Copper Pseudomonas aeruginosa Fermentation Broth and/or Copper Pseudomonas aeruginosa Metabolism Products Used for Degradation of Ochratoxin. Beijing: State Intellectual Property Office.
Liuzzi, V. C., Fanelli, F., Tristezza, M., Haidukowski, M., Picardi, E., Manzari, C., et al. (2016). Transcriptional analysis of Acinetobacter sp. neg1 capable of degrading Ochratoxin A. Front. Microbiol. 7:2162. doi: 10.3389/fmicb.2016.02162
Loi, M., Fanelli, F., Liuzzi, V. C., Logrieco, A. F., and Mulè, G. (2017). Mycotoxin biotransformation by native and commercial enzymes: present and future perspectives. Toxins (Basel) 9:111. doi: 10.3390/toxins9040111
Luz, C., Ferrer, J., Mañes, J., and Meca, G. (2018). Toxicity reduction of Ochratoxin A by lactic acid bacteria. Food Chem. Toxicol. 112, 60–66. doi: 10.1016/j.fct.2017.12.030
Madhyastha, M. S., Marquardt, R. R., and Frohlich, A. A. (1992). Hydrolysis of Ochratoxin A by the microbial activity of digesta in the gastrointestinal tract of rats. Arch. Environ. Contam. Toxicol. 23, 468–472. doi: 10.1007/bf00203811
Marin, D. E., and Taranu, I. (2015). Ochratoxin A and its effects on immunity. Toxin Rev. 34, 11–20. doi: 10.3109/15569543.2014.958757
Mateo, E. M., Medina Mateo, F., Valle-Algarra, F. M., Pardo, I., and Jiménez, M. (2010). Ochratoxin A removal in synthetic media by living and heat-inactivated cells of Oenococcus oeni isolated from wines. Food Control 21, 23–28. doi: 10.1016/j.foodcont.2009.03.012
Molnar, O., Schatzmayr, G., Fuchs, E., and Prillinger, H. (2004). Trichosporon mycotoxinivorans sp. nov., A new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 27, 661–671. doi: 10.1078/0723202042369947
Moruno, E. G., Sanlorenzo, C., Boccaccino, B., and Di Stefano, R. (2005). Treatment with yeast to reduce the concentration of Ochratoxin A in red wine. Am. J. Enol. Viticult. 56, 73–76.
Müller, H. M., Lerch, C., Müller, K., and Eggert, W. (1998). Kinetic profiles of Ochratoxin A and Ochratoxin Alpha during in vitro incubation in buffered forestomach and abomasal contents from cows. Nat. Toxins 6, 251–258. doi: 10.1002/(SICI)1522-7189(199811/12)6:6<251::AID-NT35>3.0.CO;2-P
Müller, H. M., Müller, K., and Steingass, H. (2001). Effect of feeding regime on the metabolism of Ochratoxin A during the in vitro incubation in buffered rumen fluid from cows. Arch. Anim. Nutr. 54, 265–279. doi: 10.1080/17450390109381984
Nunez, Y. P., Pueyo, E., Carrascosa, A. V., and Martínez-Rodríguez, A. J. (2008). Effects of aging and heat treatment on whole yeast cells and yeast cell walls and on adsorption of Ochratoxin A in a wine model system. J. Food Prot. 71, 1496–1499. doi: 10.4315/0362-028X-71.7.1496
Ozpinar, H., Augonyte, G., and Drochner, W. (1999). Inactivation of ochratoxin in ruminal fluid with variation of pH-value and fermentation parameters in an in vitro system. Environ. Toxicol. Pharmacol. 7, 1–9. doi: 10.1016/s1382-6689(98)00041-6
Patharajan, S., Reddy, K. R. N., Karthikeyan, V., Spadaro, D., Lore, A., Gullino, M. L., et al. (2011). Potential of yeast antagonists on invitro biodegradation of Ochratoxin A. Food Control 22, 290–296. doi: 10.1016/j.foodcont.2010.07.024
Pereyra, C. M., Cavaglieri, L. R., Keller, K. M., Chiacchera, S. M., Rosa, C. A. D. R., and Dalcero, A. M. (2015). In vitro Ochratoxin A adsorption by commercial yeast cell walls. Rev. Bras. Med. Vet. 37, 25–28.
Petchkongkaew, A., Taillandier, P., Gasaluck, P., and Lebrihi, A. (2008). Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): screening for aflatoxin B1 and Ochratoxin A detoxification. J. Appl. Microbiol. 104, 1495–1502. doi: 10.1111/j.1365-2672.2007.03700.x
Péteri, Z., Téren, J., Vágvölgyi, C., and Varga, J. (2007). Ochratoxin degradation and adsorption caused by astaxanthin-producing yeasts. Food Microbiol. 24, 205–210. doi: 10.1016/j.fm.2006.06.003
Petruzzi, L., Bevilacqua, A., Corbo, M. R., Speranza, B., Capozzi, V., and Sinigaglia, M. (2017). A Focus on Quality and Safety Traits of Saccharomyces cerevisiae Isolated from Uva di Troia Grape Variety. J. Food Sci. 82, 124–133. doi: 10.1111/1750-3841.13549
Petruzzi, L., Bevilacqua, A., Baiano, A., Beneduce, L., Corbo, M. R., and Sinigaglia, M. (2014a). In vitro removal of Ochratoxin A by two strains of Saccharomyces cerevisiae and their performances under fermentative and stressing conditions. J. Appl. Microbiol. 116, 60–70. doi: 10.1111/jam.12350
Petruzzi, L., Bevilacqua, A., Baiano, A., Beneduce, L., Corbo, M. R., and Sinigaglia, M. (2014b). Study of Saccharomyces cerevisiae W13 as a functional starter for the removal of Ochratoxin A. Food Control 35, 373–377. doi: 10.1016/j.foodcont.2013.07.033
Petruzzi, L., Bevilacqua, A., Corbo, M. R., Garofalo, C., Baiano, A., and Sinigaglia, M. (2014c). Selection of autochthonous Saccharomyces cerevisiae strains as wine starters using a polyphasic approach and Ochratoxin A removal. J. Food Prot. 77, 1168–1177. doi: 10.4315/0362-028X.JFP-13-384
Petruzzi, L., Corbo, M. R., Baiano, A., Beneduce, L., Sinigaglia, M., and Bevilacqua, A. (2015). In vivo stability of the complex Ochratoxin A – Saccharomyces cerevisiae starter strains. Food Control 50, 516–520. doi: 10.1016/j.foodcont.2014.09.042
Petruzzi, L., Corbo, M. R., Sinigaglia, M., and Bevilacqua, A. (2014d). Yeast cells as adsorbing tools to remove Ochratoxin A in a model wine. Int. J. Food Sci. Tech. 49, 936–940. doi: 10.1111/ijfs.12380
Petruzzi, L., Sinigaglia, M., Corbo, M. R., Beneduce, L., and Bevilacqua, A. (2013). Ochratoxin A removal by Saccharomyces cerevisiae strains: effect of wine-related physicochemical factors. J. Sci. Food Agric. 93, 2110–2115. doi: 10.1002/jsfa.6010
Pfliegler, W. P., Pusztahelyi, T., and Pócsi, I. (2015). Mycotoxins - prevention and decontamination by yeasts. J. Basic Microbiol. 55, 805–818. doi: 10.1002/jobm.201400833
Pfohl-Leszkowicz, A., and Manderville, R. A. (2007). Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 51, 61–99. doi: 10.1002/mnfr.200600137
Pfohl-Leszkowicz, A., and Manderville, R. A. (2012). An Update on Direct Genotoxicity as a Molecular Mechanism of Ochratoxin A Carcinogenicity. Chem. Res. Toxicol. 25, 252–262. doi: 10.1021/tx200430f
Pinotti, L., Ottoboni, M., Giromini, C., Dell'orto, V., and Cheli, F. (2016). Mycotoxin contamination in the EU feed supply chain: a focus on cereal byproducts. Toxins (Basel) 8:45. doi: 10.3390/toxins8020045
Piotrowska, M. (2012). Adsorption of Ochratoxin A by Saccharomyces cerevisiae living and non-living cells. Acta Aliment. 41, 1–7. doi: 10.1556/AAlim.2011.0006
Piotrowska, M. (2014). The Adsorption of Ochratoxin A by Lactobacillus Species. Toxins (Basel). 6:2826. doi: 10.3390/toxins6092826
Piotrowska, M., Nowak, A., and Czyzowska, A. (2013). Removal of Ochratoxin A by wine Saccharomyces cerevisiae strains. Eur. Food Res. Technol. 236, 441–447. doi: 10.1007/s00217-012-1908-3
Piotrowska, M., and Zakowska, Z. (2000). The biodegradation of Ochratoxin A in food products by lactic acid bacteria and baker's yeast. Food Biotechnol. 17, 307–310. doi: 10.1016/S0921-0423(00)80085-4
Pitout, M. J. (1969). The hydrolysis of Ochratoxin A by some proteolytic enzymes. Biochem. Pharmacol. 18, 485–491. doi: 10.1016/0006-2952(69)90224-X
Qi, X., Yang, X., Chen, S., He, X., Dweep, H., Guo, M., et al. (2014). Ochratoxin A induced early hepatotoxicity: new mechanistic insights from microRNA, mRNA and proteomic profiling studies. Sci. Rep. 4:163. doi: 10.1038/srep05163
Quintela, S., Carmen Villaran, M., Lopez De Armentia, I., and Elejalde, E. (2013). Ochratoxin A removal in wine: a review. Food Control 30, 439–445. doi: 10.1016/j.foodcont.2012.08.014
Raiola, A., Tenore, G. C., Manyes, L., Meca, G., and Ritieni, A. (2015). Risk analysis of main mycotoxins occurring in food for children: an overview. Food Chem. Toxicol. 84, 169–180. doi: 10.1016/j.fct.2015.08.023
Ringot, D., Lerzy, B., Bonhoure, J. P., Auclair, E., Oriol, E., and Larondelle, Y. (2005). Effect of temperature on in vitro Ochratoxin A biosorption onto yeast cell wall derivatives. Process Biochem. 40, 3008–3016. doi: 10.1016/j.procbio.2005.02.006
Ringot, D., Lerzy, B., Chaplain, K., Bonhoure, J.-P., Auclair, E., and Larondelle, Y. (2007). In vitro biosorption of Ochratoxin A on the yeast industry by-products: comparison of isotherm models. Bioresour. Technol. 98, 1812–1821. doi: 10.1016/j.biortech.2006.06.015
Rodrigues, I., Binder, E. M., and Schatzmayr, G. (2009). “Microorganisms and their enzymes for detoxifying mycotoxins posing a risk to livestock animals,” in Mycotoxin Prevention and Control in Agriculture, eds M. Appell, D. F. Kendra, and M. W. Trucksess (Washington, DC: American Chemical Society), 107–117.
Rodriguez, H., Reveron, I., Doria, F., Costantini, A., De Las Rivas, B., Munoz, R., et al. (2011). Degradation of Ochratoxin A by Brevibacterium species. J. Agric. Food Chem. 59, 10755–10760. doi: 10.1021/jf203061p
Russo, P., Capozzi, V., Spano, G., Corbo, M. R., Sinigaglia, M., and Bevilacqua, A. (2016). Metabolites of microbial origin with an impact on health: Ochratoxin A and biogenic amines. Front. Microbiol. 7:482. doi: 10.3389/fmicb.2016.00482
Schatzmayr, G., Heidler, D., Fuchs, E., Nitsch, S., Mohnl, M., Taubel, M., et al. (2003). Investigation of different yeast strains for the detoxification of Ochratoxin A. Mycotoxin Res. 19, 124–128. doi: 10.1007/bf02942950
Schatzmayr, G., Zehner, F., Täubel, M., Schatzmayr, D., Klimitsch, A., Loibner, A. P., et al. (2006). Microbiologicals for deactivating mycotoxins. Mol. Nutr. Food Res. 50, 543–551. doi: 10.1002/mnfr.200500181
Schwartz, G. G. (2002). Hypothesis: does Ochratoxin A cause testicular cancer? Cancer Causes Control 13, 91–100. doi: 10.1023/a:1013973715289
Shen, X. L., Zhang, B., Liang, R., Cheng, W.-H., Xu, W., Luo, Y., et al. (2014). Central role of Nix in the autophagic response to Ochratoxin A. Food Chem. Toxicol. 69, 202–209. doi: 10.1016/j.fct.2014.04.017
Shen, X. L., Zhang, Y., Xu, W., Liang, R., Zheng, J., Luo, Y., et al. (2013). An iTRAQ-based mitoproteomics approach for profiling the nephrotoxicity mechanisms of Ochratoxin A in HEK 293 cells. J. Proteomics 78, 398–415. doi: 10.1016/j.jprot.2012.10.010
Sherazi, S. T. H., Shar, Z. H., Sumbal, G. A., Tan, E. T., Bhanger, M. I., Kara, H., et al. (2015). Occurrence of Ochratoxin A in poultry feeds and feed ingredients from Pakistan. Mycotoxin Res. 31, 1–7. doi: 10.1007/s12550-014-0216-0
Shi, L., Liang, Z., Li, J., Hao, J., Xu, Y., Huang, K., et al. (2014). Ochratoxin A biocontrol and biodegradation by Bacillus subtilis CW 14. J. Sci. Food Agric. 94, 1879–1885. doi: 10.1002/jsfa.6507
Shi, L., Liang, Z., Xu, S., Zheng, H., and Huang, K. (2013). Adsorption and degradation of Ochratoxin A by Bacillus licheniformis Sl-1. J. Agric. Biotechnol. 21, 1420–1425. doi: 10.3969/j.issn.1674-7968.2013.12.003
Slizewska, K., and Piotrowska, M. (2014). Reduction of Ochratoxin A in chicken feed using probiotic. Ann. Agric. Environ. Med. 21, 676–680. doi: 10.5604/12321966.1129913
Spano, G., and Capozzi, V. (2011). Food Microbial Biodiversity and “Microbes of Protected Origin”. Front. Microbiol. 2:237. doi: 10.3389/fmicb.2011.00237
Stander, M. A., Bornscheuer, U. T., Henke, E., and Steyn, P. S. (2000). Screening of commercial hydrolases for the degradation of Ochratoxin A. J. Agric. Food Chem. 48, 5736–5739. doi: 10.1021/jf000413j
Streit, E., Naehrer, K., Rodrigues, I., and Schatzmayr, G. (2013). Mycotoxin occurrence in feed and feed raw materials worldwide: long-term analysis with special focus on Europe and Asia. J. Sci. Food Agric. 93, 2892–2899. doi: 10.1002/jsfa.6225
Styriak, I., Conková, E., Kmec, V., Böhm, J., and Razzazi, E. (2001). The use of yeast for microbial degradation of some selected mycotoxins. Mycotoxin Res. 17, 24–27. doi: 10.1007/bf03036705
Sun, X. D., Su, P., and Shan, H. (2017). Mycotoxin Contamination of Maize in China. Compr. Rev. Food Sci. F. 16, 835–849. doi: 10.1111/1541-4337.12286
Susca, A., Proctor, R. H., Morelli, M., Haidukowski, M., Gallo, A., Logrieco, A. F., et al. (2016). Variation in fumonisin and ochratoxin production associated with differences in biosynthetic gene content in Aspergillus niger and A. welwitschiae Isolates from Multiple Crop and Geographic Origins. Front. Microbiol. 7:1412. doi: 10.3389/fmicb.2016.01412
Taheur, F. B., Fedhila, K., Chaieb, K., Kouidhi, B., Bakhrouf, A., and Abrunhosa, L. (2017). Adsorption of aflatoxin B1, zearalenone and Ochratoxin A by microorganisms isolated from Kefir grains. Int. J. Food Microbiol. 251, 1–7. doi: 10.1016/j.ijfoodmicro.2017.03.021
Upadhaya, S. D., Song, J. Y., Park, M. A., Seo, J. K., Yang, L., Lee, C. H., et al. (2012). Isolation, screening and identification of swine gut microbiota with Ochratoxin A biodegradation ability. Asian Austral. J. Anim. 25, 114–121. doi: 10.5713/ajas.2011.11104
Upadhaya, S. D., Yang, L., Seo, J. K., Kim, M. H., Lee, C.-K., Lee, C. H., et al. (2011). Effect of feed types on Ochratoxin A disappearance in goat rumen fluid. Asian Austral. J. Anim. 24, 198–205. doi: 10.5713/ajas.2011.10318
Valero, A., Sanchis, V., Ramos, A. J., and Marín, S. (2008). Brief in vitro study on Botrytis cinerea and Aspergillus carbonarius regarding growth and Ochratoxin A. Lett. Appl. Microbiol. 47, 327–332. doi: 10.1111/j.1472-765X.2008.02434.x
Van Der Merwe, K. J., Steyn, P. S., Fourie, L., Scott, D. B., and Theron, J. J. (1965). Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature 205, 1112–1113. doi: 10.1038/2051112a0
Var, I., Erginkaya, Z., and Kabak, B. (2009). Reduction of Ochratoxin A Levels in White Wine by Yeast Treatments. J. Ins. Brewing 115, 30–34. doi: 10.1002/j.2050-0416.2009.tb00341.x
Varga, J., Péteri, Z., Tábori, K., Téren, J., and Vágvölgyi, C. (2005). Degradation of Ochratoxin A and other mycotoxins by Rhizopus isolates. Int. J. Food Microbiol. 99, 321–328. doi: 10.1016/j.ijfoodmicro.2004.10.034
Varga, J., Rigó, K., and Téren, J. (2000). Degradation of Ochratoxin A by Aspergillus species. Int. J. Food Microbiol. 59, 1–7. doi: 10.1016/s0168-1605(00)00230-0
Wang, J., Zhang, H., and Yang, Q. (2014). Screening and identification of microorganisms for Ochratoxin A degradation. Food Sci. 35, 154–158. doi: 10.7506/spkx1002-6630-201421030
Wang, Y., Peng, X., Yang, Z., Zhao, W., Xu, W., Hao, J., et al. (2017). iTRAQ Mitoproteome analysis reveals mechanisms of programmed cell death in Arabidopsis thaliana induced by Ochratoxin A. Toxins (Basel) 9:167. doi: 10.3390/toxins9050167
Wang, Y., Wang, L., Liu, F., Wang, Q., Selvaraj, J., Xing, F., et al. (2016). Ochratoxin A producing fungi, biosynthetic pathway and regulatory mechanisms. Toxins (Basel) 8:83. doi: 10.3390/toxins8030083
Wegst, W., and Lingens, F. (1983). Bacterial degradation of Ochratoxin A. FEMS Microbiol. Lett. 17, 341–344. doi: 10.1111/j.1574-6968.1983.tb00433.x
Wu, Q., Dohnal, V., Huang, L., Kuca, K., Wang, X., Chen, G., et al. (2011). Metabolic Pathways of Ochratoxin A. Curr. Drug Metab. 12, 1–10. doi: 10.2174/138920011794520026
Xiao, H., Marquardt, R. R., Frohlich, A. A., Phillips, G. D., and Vitti, T. G. (1991). Effect of a hay and a grain diet on the bioavailability of Ochratoxin A in the rumen of sheep. J. Anim. Sci. 69, 3715–3723. doi: 10.2527/1991.6993715x
Xiong, K., Wang, X.-L., Zhi, H.-W., Sun, B.-G., and Li, X.-T. (2017). Identification and safety evaluation of a product from the biodegradation of Ochratoxin A by an Aspergillus strain. J. Sci. Food Agric. 97, 434–443. doi: 10.1002/jsfa.7742
Yang, Q., Wang, J., Zhang, H., Li, C., and Zhang, X. (2016). Ochratoxin A is degraded by Yarrowia lipolytica and generates non-toxic degradation products. World Mycotoxin J. 9, 269–278. doi: 10.3920/wmj2015.1911
Yu, S., Poulsen, C. H., Dalsgaard, S., Wang, H., and Nikolaev, I. (2015). Food Additive Comprising an Amidase for Detoxifying Ochratoxin. Beijing: State Intellectual Property Office.
Zhang, B., Shen, X. L., Liang, R., Li, Y., Huang, K., Zhao, C., et al. (2014). Protective role of the mitochondrial Lon protease 1 in Ochratoxin A-induced cytotoxicity in HEK293 cells. J. Proteomics 101, 154–168. doi: 10.1016/j.jprot.2014.02.017
Zhang, H. H., Wang, Y., Zhao, C., Wang, J., and Zhang, X. L. (2017). Biodegradation of Ochratoxin A by Alcaligenes faecalis isolated from soil. J. Appl. Microbiol. 123, 661–668. doi: 10.1111/jam.13537
Zhao, T., Shen, X. L., Chen, W., Liao, X., Yang, J., Wang, Y., et al. (2017). Advances in research of nephrotoxicity and toxic antagonism of Ochratoxin A. Toxin Rev. 36, 39–44. doi: 10.1080/15569543.2016.1243560
Zheng, J., Zhang, Y., Xu, W., Luo, Y., Hao, J., Shen, X. L., et al. (2013). Zinc protects HepG2 cells against the oxidative damage and DNA damage induced by Ochratoxin A. Toxicol. Appl. Pharmacol. 268, 123–131. doi: 10.1016/j.taap.2013.01.021
Keywords: Ochratoxin A, biodegradation, adsorption, bacteria, filamentous fungi, yeast
Citation: Chen W, Li C, Zhang B, Zhou Z, Shen Y, Liao X, Yang J, Wang Y, Li X, Li Y and Shen XL (2018) Advances in Biodetoxification of Ochratoxin A-A Review of the Past Five Decades. Front. Microbiol. 9:1386. doi: 10.3389/fmicb.2018.01386
Received: 06 March 2018; Accepted: 06 June 2018;
Published: 26 June 2018.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Lucilla Iacumin, Università degli Studi di Udine, ItalyFrancesco Grieco, Istituto di Scienze delle Produzioni Alimentari (ISPA), Italy
Copyright © 2018 Chen, Li, Zhang, Zhou, Shen, Liao, Yang, Wang, Li, Li and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao L. Shen, xiaolishen1983@163.com
†These authors have contributed equally to this work.
 Wenying Chen1,2†
Wenying Chen1,2† Boyang Zhang
Boyang Zhang Xiao L. Shen
Xiao L. Shen