- 1Pathogen Genomics Center, National Institute of Infectious Diseases, Tokyo, Japan
- 2Antimicrobial Resistance Research Center, National Institute of Infectious Diseases, Higashimurayama, Japan
- 3The Public Health Institute of Kochi Prefecture, Kōchi City, Japan
A novel species of carbapenemase-producing Enterobacteriaceae (CPE) was isolated from a patient diagnosed with sigmoid colon diverticulitis. At first, laboratory testing suggested it was Klebsiella oxytoca or Pantoea sp.; however, a complete genome sequence of the isolate, MRY16-398, revealed that it could be novel species, most similar to [Kluyvera] intestini, of which taxonomic nomenclature is still under discussion. Orthologous conserved gene analysis among 42 related bacterial strains indicated that MRY16-398 was classified as the newly proposed genus Metakosakonia. Further, MRY16-398 was found to harbor the blaIMP-6 gene-positive class 1 integron (In722) in plasmid pMRY16-398_2 (IncN replicon, 47.4 kb in size). This finding implies that rare and opportunistic bacteria could be potential infectious agents. In conclusion, our results highlight the need for continuous monitoring for CPE even in nonpathogenic bacteria in the nosocomial environment.
Introduction
Antimicrobial resistance (AMR) is a global issue linked to increased and often unrestricted antibiotic use in the clinical settings, which leads to the dissemination of carbapenem-resistant Enterobacteriaceae (CRE) in health care facilities (World Health Organization, 2017). Carbapenemases comprise three of the four Ambler classes as follows: Class A (Klebsiella pneumoniae carbapenemases, KPC, some variants of Guiana extended-spectrum β-lactamases, GES), Class B (metallo-β-lactamases, MBL including New Delhi metallo-β-lactamases, NDM, Verona integron-encoded metallo-β-lactamases, VIM, and imipenemase, IMP), and Class D (OXA-48-like carbapenemases) (Logan and Weinstein, 2017). These carbapenemase-producing Enterobacteriaceae (CPE) have the potential to facilitate the widespread transmission of antimicrobial resistance genes (ARGs) via mobile genetic elements through processes including natural competence, transformation, and plasmid transconjugation that can occur in any environment (Kelly et al., 2017; Rozwandowicz et al., 2018).
The widespread detection of CPE is an emerging issue with potentially serious public health implications; further, the distribution of the most common carbapenemase genes in Enterobacteriaceae occurs in a country- and region-specific manner (Logan and Weinstein, 2017). In Japan, IMP is the most predominant type of carbapenemase among clinical CPE isolates (Koyano et al., 2013; Ohno et al., 2017; Yamamoto et al., 2017). VIM, OXA-48-like, KPC, and NDM carbapenemases are detected at low frequencies in Japan, whereas KPC and NDM are predominant in China and OXA-48-like and KPC are the predominant types of carbapenemases in Europe and United States, respectively.
Most CRE/CPE infections occur in hospitals, with major outbreaks at long-term care facilities and affecting patients with severe medical conditions under long stays for clinical management (Grabowski et al., 2017). There are a number of factors that predispose individuals to infections by CRE and other multi-drug resistant Enterobacteriaceae, including extended-spectrum β-lactamase (ESBL)-producers. Indeed, healthy carriers of CTX-M-type ESBL-harboring bacteria represent major public health concerns, because the carriage rates are on the rise, particularly in South East Asia and Eastern Mediterranean regions. Further, carriers from these regions could potentially spread these bacteria to other communities (Woerther et al., 2013).
Exposure to AMR bacteria can cause serious infections in patients with risk factors such as empirical antimicrobials, advanced age, immune-suppression, admission to the intensive care unit, mechanical ventilation, transplantation, and prolonged hospital stay (Gasink et al., 2009). Early intervention, through the administration of effective antimicrobials to such high-risk group patients, must be achieved to prevent death. In addition, a recent systematic review identified a prevalence of 0–29.5% for community-associated CRE, suggesting that the early detection of CRE-carriers among hospitalized patients upon admission to long-term care facilities might help to prevent nosocomial outbreaks and control the limited distribution of such emerging public health threats (Kelly et al., 2017).
Generally, Klebsiella, Escherichia coli, Enterobacter, and Citrobacter have been reported as the main contributors to the nosocomial transmission of CPE (Hrabak et al., 2014; Goodman et al., 2016; Kwong et al., 2018). Other opportunistic pathogens among Enterobacteriaceae species can acquire carbapenemase genes through plasmid transmission from main CPE contributors. Kluyvera is a group of gram-negative rod-shaped bacteria and is a member of the Enterobacteriaceae family; the genus contains four species, namely Kluyvera ascorbata, Kluyvera cryocrescens, Kluyvera georgiana, and Kluyvera intermedia, which have all been found in humans (Farmer et al., 1981). K. ascorbata and K. cryocrescens were reported as potential pathogens that are associated with sepsis, bacteremia, catheter-associated urinary tract infections, pyelonephritis, and intraabdominal symptoms in immunocompromised hosts (Karadag Oncel et al., 2015; Yoshino et al., 2016).
Here, we report a novel IMP-6-producing isolate of Metakosakonia sp., namely strain MRY16-398, from a clinical specimen (ascites), and determined the genomic features of this carbapenemase-producing species.
Materials and Methods
Ethics Approval
The study protocol was approved by the ethics committee of the National Institute of Infectious Diseases in Japan (Approval No. 642, 11/Dec/2015). It was conducted according to the principles of the Declaration of Helsinki, in compliance with the Law Concerning the Prevention of Infections and Medical Care for Patients of Infections of Japan; the ethical committee waived the need for written consent regarding the research of bacteria isolates; the personal data related to the clinical information were anonymized, and we do not request written consent for all patients suffering from bacterial infections.
Bacterial Strains and Identification
Upon admitting a patient complaining of acute abdominal pain, abdominal computed tomography (CT) scanning showed a diverticulum in the descending and sigmoid colon, resulting in the diagnosis of sigmoid colon diverticulitis. A summary of laboratory data for the patient is shown in Supplementary Table S1. Empiric antimicrobial treatment with cefmetazole (0.5 g twice per day) was administered, and the volume of ascites was decreasing at 5 days from onset.
The Metakosakonia sp. MRY16-398 strain was isolated from the ascites of a patient with sigmoid colon diverticulitis in 2015 in Japan. The isolate was identified as Klebsiella oxytoca at the hospital microbiology laboratory using BD Phoenix (Becton Dickinson) with low reliability. Further phenotypic tests were performed using API 20E (bioMérieux) and Lysine-Indole-Motility Medium (Nissui, Tokyo Japan). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)-based identification was conducted with a MicroFlex LT mass spectrometer (Bruker Daltonik), and analyzed using MALDI Biotyper software (Bruker Daltonik).
Antimicrobial Susceptibility and CPE Screening Tests
Antimicrobial susceptibility was investigated by broth microdilution using the MicroScan Neg MIC 1J panel (Beckman Coulter) and Etest (bioMérieux) according to manufacturers’ instructions (Clsi, 2018). Boronic acid, clavulanic acid, and sodium mercaptoacetic acid (SMA) were used as inhibitors for double-disk synergy tests (DDSTs) to identify AmpC-types and KPC-types, as well as extended-spectrum and metallo-β-lactamases, respectively. Carbapenemase production was assessed by performing a Carba NP test, as described previously (Nordmann et al., 2012). PCR testing was subsequently performed for potential CPE using a specific primer-pair for the following types of β-lactamase-encoding genes: blaIMP (Shibata et al., 2003), blaV IM (Shibata et al., 2003), blaOXA-48-like (Poirel et al., 2004), blaKPC (Bradford et al., 2004), and blaNDM (Segawa et al., 2017).
Plasmid and Chromosome DNA Analysis With Short-Read Sequencing
Plasmid DNA was separated from chromosomal DNA by S1 nuclease-digestion followed by pulsed-field gel electrophoresis. Visible plasmid DNA and chromosomal DNA bands were extracted from the gel and purified using the ZR-96 Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA, United States). A DNA sequencing library was prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, United States) and was sequenced using an Illumina MiSeq and NextSeq 500 for plasmids and chromosomes, respectively. Sequencing reads (plasmid: 2 × 300-mer, 140 × median coverage; chromosome: 2 × 150-mer, 99 × median coverage) were assembled into contigs using the A5-MiSeq pipeline (Coil et al., 2015). Plasmid replicon typing was performed using the curated PlasmidFinder database at the CGE website1 (Carattoli et al., 2014).
Whole-Genome Sequence (WGS) Analysis With Long-Read Sequencing
Genomic DNA from the isolated strain was purified by collecting cells from a 5-mL overnight culture grown in TSB broth. The cell pellet was resuspended in 500 μL of TE10 [10 mM tris (pH 8.0) and 10 mM EDTA] supplemented with 500 μL phenol/chloroform, and the cells were subsequently lysed by bead-beating for 10 min in ZR BashingBead lysis tubes (Zymo Research, Irvine, CA, United States) attached to a vortex adapter (MO BIO Laboratories, QIAGEN, Carlsbad, CA, United States). After centrifugation at 10,000 × g for 5 min; the upper phase was further purified using a Qiagen DNA purification kit (Qiagen, Germany).
The complete genome sequence of the strain was determined using a PacBio RSII sequencer for long-read sequencing (SMRT cell v3 using P6C4 chemistry; insert size, approximately 10 kb). Purified genomic DNA (∼2.0 μg) was used to prepare a SMRTbell library using a SMRTbell template prep kit 1.0 (PacBio, Menlo Park, CA, United States) according to manufacturer’s instructions. The obtained raw polymerase reads were analyzed using the HGAP v3.0 pipeline based on Celera de novo assembler and Quiver polishing scripts (Chin et al., 2013).
De novo assembly was performed using HGAP 4 of SMRT Link Analysis v. 4.0.0.190159 and circulator version 1.5.3 (Chin et al., 2013). Error correction of tentative complete circular sequences was performed using Pilon version 1.18 with Illumina short reads (Walker et al., 2014). Annotation was performed using DFAST (Tanizawa et al., 2018) with databases as follows: DFAST default database, ResFinder database (Zankari et al., 2012), Bacterial Antimicrobial Resistance Reference Gene Database (PRJNA313047), and Virulence Factors Database (Chen et al., 2012). Circular representations of complete plasmid sequences were visualized using GView server (Petkau et al., 2010).
Comparative Genome Sequence Analysis
All publicly available draft genome sequences were searched based on 16S rRNA gene homology, comparing them to that of the MRY16-398 strain, and 41 entries were retrieved from the NCBI genome database (see Supplementary Table S2). Among those 42 strains, orthologous core-gene sets were extracted using a nucleotide homology search with a threshold ≥80%, resulting in the identification of 479 core-gene sets (see Supplementary Table S2). Using these core-gene sets, core-gene phylogeny was generated using the maximum-likelihood phylogenetic method with FastTree v2.1.10 (Price et al., 2010). To construct a pairwise amino acid homology distance matrix, all amino acid sequences were compared pairwise, against each other, for each genome using the BLASTP program, which was followed by the calculation of average identity scores and standard deviations (Supplementary Table S3).
Nucleotide Sequence Accession Numbers
The complete, annotated genomic sequence of Metakosakonia sp. MRY16-398 was deposited in a public database (accession numbers: chromosome, AP018756; pMRY16-398_1, AP018757; pMRY16-398_2, AP018758). The short- and long-read sequences for DNA-Seq were deposited in the DNA Data Bank of Japan (BioProject PRJDB7098, BioSample SSUB009772, DRA accession DRA007011).
Results
Bacterial Identification of Metakosakonia sp. MRY16-398
A potential CPE, designated as strain MRY16-398, was isolated from the ascites after abdominocentesis. The isolate was identified as Klebsiella oxytoca at the hospital laboratory using BD Phoenix (Becton Dickinson) with low reliability, whereas API20E testing indicated the isolate should be a Pantoea sp. instead of K. oxytoca. This isolate was negative for lysine decarboxylate activity and showed weak motility, which indicated that the isolate was not Klebsiella. MALDI-TOF MS did not result in any bacterial species with a score higher than 2.000, which secures genus and probable species identification. The highest score value was 1.885 for Klebsiella aerogenes, followed by 1.789 for K. oxytoca.
Generally, 16S-rRNA gene sequencing is one of first tools used to determine the correct bacterial species of such novel CPE isolates, and thus we considered that WGS would be a more straightforward approach to characterize the species and plasmids involved in AMR.
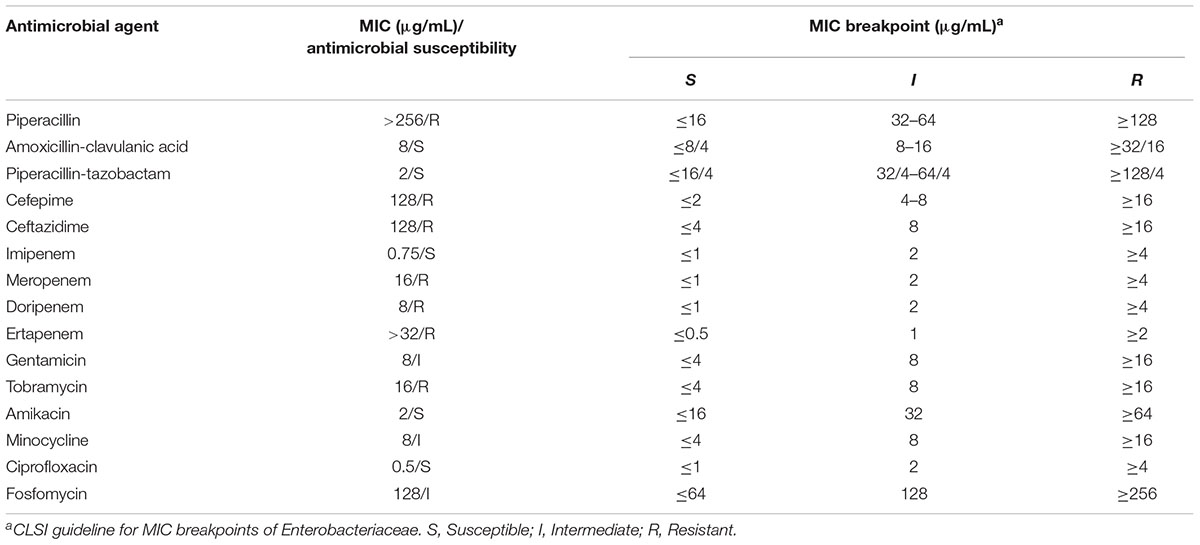
The MRY16-398 isolate was observed to harbor blaIMP-6, exhibited resistance to meropenem, and was positive based on the Carba NP test and DDST using SMA. Further antimicrobial susceptibility testing indicated that MRY16-398 exhibited remarkably reduced susceptibility to most β-lactam antibiotics (Table 1).
Whole-Genome Sequence Analysis of Metakosakonia sp. MRY16-398
Basic information regarding the complete chromosome and plasmid sequences of Metakosakonia sp. MRY16-398 is shown in Table 2. The strain possessed two plasmids, and pMRY16-398_2 was determined to be an IncN replicon plasmid, harboring multiple AMR-encoding genes including the blaIMP-6 carbapenemase-encoding gene (Table 2). The IMP-6 metallo-β-lactamase is an IMP variant with a S214G amino acid substitution in the catalytic domain of IMP-1, resulting in significantly diminished enzymatic activity toward imipenem but not meropenem (Oelschlaeger et al., 2005). Thus, MRY16-398 showed susceptibility to imipenem, but resistance to other carbapenems (Table 1). The aacA4′-3 gene encoding aminoglycoside-3′′-adenylyltransferase, and the aadA2 gene encoding streptomycin 3′′-adenylyltransferase are involved in resistance to aminoglycosides. The tet(A) gene could be involved in reduced susceptibility to minocycline. Comparative analyses of the MRY16-398 genome sequence including the pMRY16-398_2 plasmid are described in the following section.
Orthologous Gene Phylogenetic Analysis of Metakosakonia sp. MRY16-398
To determine the potential bacterial species of the MRY16-398 strain, we performed orthologous gene phylogenetic analysis using 41 publicly available bacterial genome sequences (on 2017/03/14), including draft genomes (see the strain list in Supplementary Table S2). Among those 42 strains including MRY16-398, 479 orthologous gene sets were extracted at ≥80% nucleotide identity, and phylogeny and matrix distance clearly suggested that MRY16-398 was closely related to the bacterial species [Kluyvera] intestini str. GT-16 (Tetz et al., 2017), which was isolated from the stomach of a patient with gastric cancer (Figure 1). Recently, the taxonomic nomenclature for [Kluyvera] intestini str. GT-16 has been re-classified into a new proposed genera, namely Metakosakonia, which includes M. massiliensis JC163 (Alnajar and Gupta, 2017). As well as the proposal, this study demonstrated GT-16 strain shows the closest lineage to Metakosakonia, and having a distinct lineage from the main Kluyvera species (K. georgiana and K. intermedia) (Figure 1). Thus, MRY16-398 was found to be a clearly distinct lineage from Kluyvera, Pantoea, and other well-characterized genera of the Enterobacteriaceae family (Figure 1), indicating that this clinical isolate is a novel species. Here, we tentatively classified MRY16-398 as Metakosakonia sp.
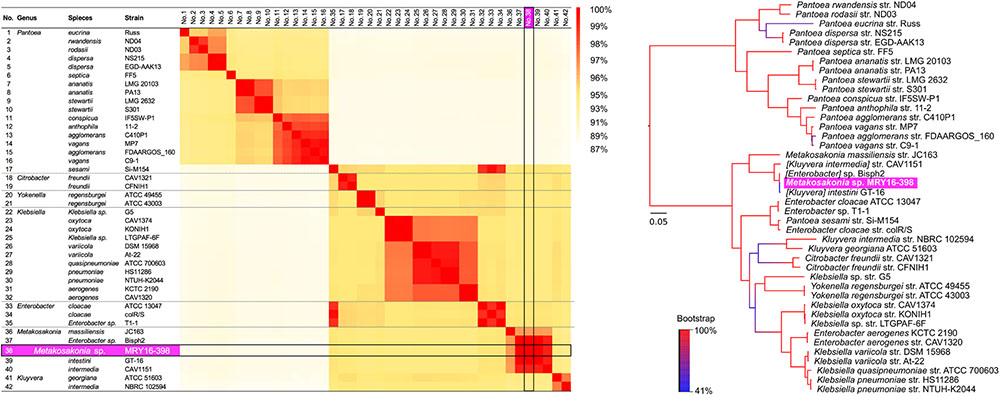
FIGURE 1. Pairwise homology sequence analysis using 479 core-gene sets from 42 strains related to MRY16-398. Maximum-likelihood phylogenetic analysis with 1,000× bootstrapping and the generation of a pairwise homology matrix were performed for the core-gene set listed in Supplementary Table S2. The MRY16-398 strain is highlighted with a bright purple background. The phylogeny and matrix distance clearly suggested that MRY16-398 is closely related to the bacterial species [Kluyvera] intestini str. GT-16 (Tetz et al., 2017), and distinct from other Enterobacteriaceae. Detailed homology % values for the matrix distances can be seen in Supplementary Table S3.
Structural Comparison of pMRY16-398_2-Associated IncN Plasmids
S1-PFGE suggested that MRY16-398 carries two plasmids (Figure 2A), and the size of both plasmids corresponded to sequencing results as well as whole genome sequencing (Table 2). An analysis of conserved genes in the pMRY16-398_2 plasmid indicated that horizontally acquired AMR genes [class 1 integron, blaCTX-M-2, and tet(A)] are variable in each plasmid, although IncN backbone regions remained well conserved (Figure 2B). The class 1 integron has been classified as In722 (intI1, aacA4’-3, blaIMP-6, aadA2, and gcISKpn22) in the INTEGRALL database2 (Moura et al., 2009). pMRY16-398_2 shared an identical ARG profile and organization with pKPI-6 from Klebsiella pneumoniae KPI-6 (Figure 2B).
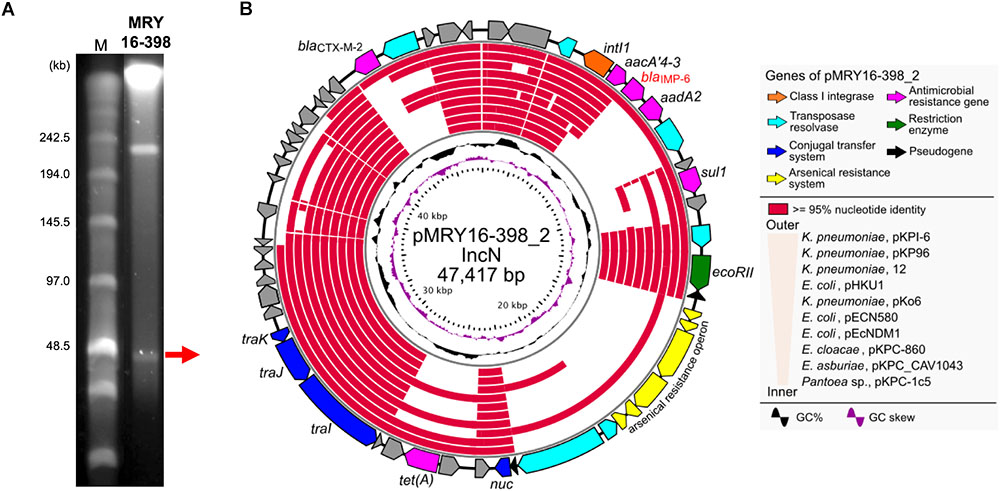
FIGURE 2. Representation of conserved gene analysis of a plasmid from Metakosakonia sp. MRY16-398. (A) Plasmids were identified by pulsed-field gel electrophoresis using an S1 nuclease-treated genomic DNA plug. (B) Circular representation of the plasmid pMRY16-398_2 carrying blaIMP-6, and conserved gene analysis, with comparative information for other indicated similar plasmids (Supplementary Table S4). From inward, slot 1, GC skew; slot 2, GC content; slot 3 to 12, source of IncN plasmids (see Supplementary Table S4), slot 13 and 14, open reading frames.
Pairwise alignment clearly showed that some genes involved in the conjugal transfer system have been excised and replaced with arsenical resistance proteins (Ars system) (Diorio et al., 1995) via an IS6100-mediated homologous recombination event (Figure 3). A mating transconjugation experiment to recipient E. coli showed negative plasmid transmission with pMRY16-398_2, although positive transmission was observed with a certain IncN plasmid harboring a full set of tra genes (data not shown). In addition to multiple AMR genes, pMRY16-398_2 appears to have lost its self-conjugation transfer ability to other bacteria, whereas it acquired arsenic resistance. This likely led to an increase in the persistence and fitness of the novel bacterial species, which is an opportunistic pathogen, in the presence of high concentrations of disinfectants in the hospital environment.
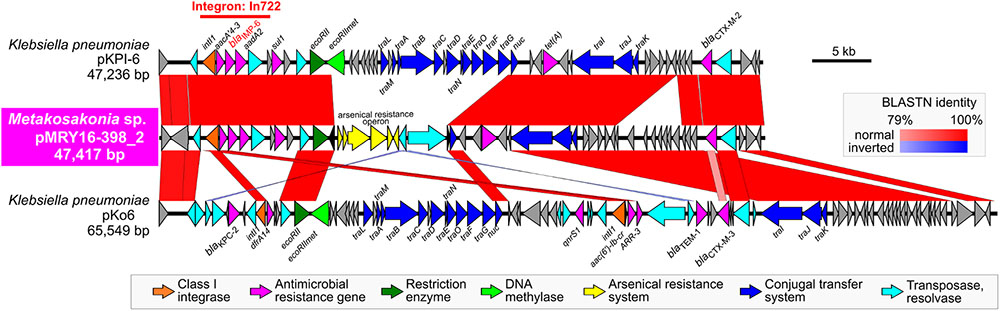
FIGURE 3. Structural comparison of the blaIMP-6-positive IncN plasmids. pMRY16-398_2 was aligned with IncN plasmid pKPI-6 (GenBank ID: AB616660) from Klebsiella pneumoniae KPI-6 isolated in 2007 in Japan. Homologous regions between the two are shown as red boxes with the color gradient corresponding to % homology. These plasmids carry the class 1 integron, In722 (intI1, aacA4’-3, blaIMP-6, aadA2, and gcISKpn22), classified in the INTEGRALL database (http://integrall.bio.ua.pt/) (Moura et al., 2009), and additional blaCTX-M-2 and tet (A).
Additional Potential AMR Genes
A search for ARGs revealed an additional potential class A β-lactamase (MRY16398_50310), with 76% amino acid similarity to the TEM-1A variant, in the chromosomal DNA of MRY16-398 (Figure 4). A maximum-likelihood phylogeny among TEM-1A-related class A β-lactamases suggested that MRY16398_50310 is closely related to those of Kluyvera spp.
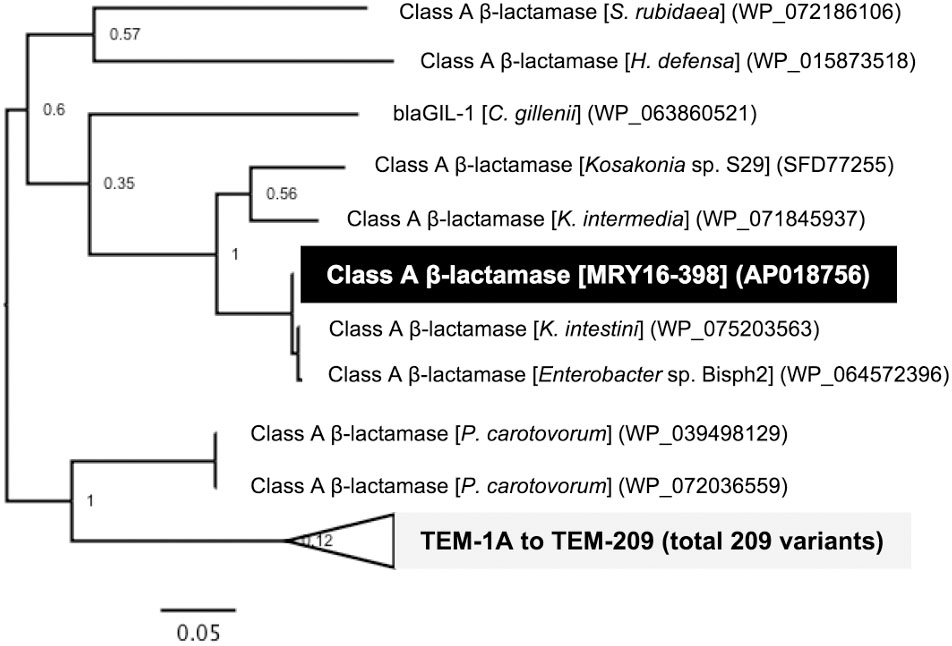
FIGURE 4. Class A β-lactamase-encoding genes in the chromosome of Metakosakonia sp. MRY16-398. A maximum-likelihood phylogeny among TEM-1A-related class A β-lactamases suggesting that MRY16398_50310 encodes a possible class A β-lactamase. Bootstrapping values are shown at the nodes.
Discussion
In this study, we isolated an IMP-6-producing novel Enterobacteriaceae species, Metakosakonia sp. strain MRY16-398, from the ascites of a diverticulitis patient. Nosocomial CPE outbreaks are generally caused by virulent pathogens; however, avirulent bacteria can cause opportunistic infection as apparent pathogens upon acquiring a notable resistance determinant. Such rare cases of avirulent or novel bacteria species have are not often documented as case reports, because conventional testing for bacterial identification are not always correct for novel species, as shown in this study. WGS comprised a straightforward approach to characterize the overall features of this isolate and its plasmids involved in AMR, and this genome sequence will be helpful for further characterization of infections caused by Metakosakonia sp.
A few studies related to the Metakosakonia genus have been reported thus far, and the most genetically related genus Kluyvera represents an informative reference for further discussion. Kluyvera spp. strains have been reported as potential pathogens in immunocompromised hosts; in addition, the Kluyvera genus is one source of CTX-M genes, which are thought to be the most common and important extended-spectrum β-lactamase-encoding genes (Humeniuk et al., 2002). For example, KLUA-producing Kluyvera ascorbata can survive for a long time in environments such as sewage and the human gut and promote drug resistance-associated gene transfer (Farmer et al., 1981). Based on a report of AMR in Kluyvera-related species, blaGES-5 positive, carbapenem-resistant K. intermedia were isolated from a hospital environment (Ribeiro et al., 2014). Further, KPC-2-producing K. ascorbata has been reported in a case of biliary tract infection (Wang et al., 2018). A K. ascorbata isolate positive for the colistin resistance gene, mcr-1, was identified from hospital sewage in China (Zhao and Zong, 2016). Such opportunistic pathogens including Kluyvera represent important multi-drug resistant bacteria in clinical settings and other environmental sources.
Here, we isolated a novel species, Metakosakonia sp. MRY16-398, and revealed the horizontal acquisition of the blaIMP-6 plasmid in this novel species that is rarely associated with the clinical settings. Such novel opportunistic pathogens might act as a potential reservoir/source of clinically relevant antibiotic resistance genes. In conclusion, these findings highlight the fact that bacterial identification is a crucial primary step when an isolate exhibits markedly reduced susceptibility as CPE. Moreover, continuous and comprehensive monitoring including WGS should be conducted for the detection of CPE even in nonpathogenic bacteria isolated from the clinical settings.
Data Availability Statement
The complete, annotated genomic sequence of [Kluyvera] intestini MRY16-398 was deposited in a public database (Accession Nos. chromosome, AP018756; pMRY16-398_1, AP018757; pMRY16-398_2, AP018758). The short- and long-read sequences for DNA-Seq were deposited in the DNA Data Bank of Japan (BioProject PRJDB7098, BioSample SSUB009772, DRA accession DRA007011).
Author Contributions
TT and AT contributed to the isolation of the IMP-6 positive Metakosakonia sp. strain MRY16-398. MM performed S1-PFGE analysis to detect individual plasmids. MM and SS performed antimicrobial susceptibility testing and DNA preparation for whole genome sequencing. TS and MK performed genome sequencing and the comparative genome analysis. MH performed mating transconjugation experiments. MK wrote the manuscript.
Funding
This work was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development, AMED (Grant Nos. JP17fk0108121, JP18fk0108048, and JP18fk0108019). The funding agencies had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Kumiko Kai for technical assistance with S1-PFGE analysis, and also Miho Nishio and Norimi Kishi for technical assistance with NGS experiments. We would also like to thank Editage (www.editage.com) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02853/full#supplementary-material
Footnotes
References
Alnajar, S., and Gupta, R. S. (2017). Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect. Genet. Evol. 54, 108–127. doi: 10.1016/j.meegid.2017.06.024
Bradford, P. A., Bratu, S., Urban, C., Visalli, M., Mariano, N., Landman, D., et al. (2004). Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 39, 55–60. doi: 10.1086/421495
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Xiong, Z., Sun, L., Yang, J., and Jin, Q. (2012). VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40, D641–D645. doi: 10.1093/nar/gkr989
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Clsi. (2018). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Eighth Informational Supplement M100–S28. Wayne, PA: CLSI.
Coil, D., Jospin, G., and Darling, A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31, 587–589. doi: 10.1093/bioinformatics/btu661
Diorio, C., Cai, J., Marmor, J., Shinder, R., and DuBow, M. S. (1995). An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J. Bacteriol. 177, 2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995
Farmer, J. J. III, Fanning, G. R., Huntley-Carter, G. P., Holmes, B., Hickman, F. W., Richard, C., et al. (1981). Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp. nov. and Kluyvera cryocrescens sp. nov. in clinical specimens. J. Clin. Microbiol. 13, 919–933.
Gasink, L. B., Edelstein, P. H., Lautenbach, E., Synnestvedt, M., and Fishman, N. O. (2009). Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 30, 1180–1185. doi: 10.1086/648451
Goodman, K. E., Simner, P. J., Tamma, P. D., and Milstone, A. M. (2016). Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev. Anti Infect. Ther. 14, 95–108. doi: 10.1586/14787210.2016.1106940
Grabowski, M. E., Kang, H., Wells, K. M., Sifri, C. D., Mathers, A. J., and Lobo, J. M. (2017). Provider role in transmission of carbapenem-resistant Enterobacteriaceae. Infect. Control Hosp. Epidemiol. 38, 1329–1334. doi: 10.1017/ice.2017.216
Hrabak, J., Chudackova, E., and Papagiannitsis, C. C. (2014). Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin. Microbiol. Infect 20, 839–853. doi: 10.1111/1469-0691.12678
Humeniuk, C., Arlet, G., Gautier, V., Grimont, P., Labia, R., and Philippon, A. (2002). Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46, 3045–3049. doi: 10.1128/AAC.46.9.3045-3049.2002
Karadag Oncel, E., Ozsurekci, Y., Akyon, Y., Gur, D., Cengiz, A. B., and Kara, A. (2015). Kluyvera ascorbata infections in children: a case series. Turk. Pediatri. Ars. 50, 123–128. doi: 10.5152/tpa.2015.923
Kelly, A. M., Mathema, B., and Larson, E. L. (2017). Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int. J. Antimicrob. Agents 50, 127–134. doi: 10.1016/j.ijantimicag.2017.03.012
Koyano, S., Saito, R., Nagai, R., Tatsuno, K., Okugawa, S., Okamura, N., et al. (2013). Molecular characterization of carbapenemase-producing clinical isolates of Enterobacteriaceae in a teaching hospital. Jpn. J. Med. Microbiol. 62(Pt 3), 446–450. doi: 10.1099/jmm.0.050708-0
Kwong, J. C., Lane, C. R., Romanes, F., Goncalves da Silva, A., Easton, M., Cronin, K., et al. (2018). Translating genomics into practice for real-time surveillance and response to carbapenemase-producing Enterobacteriaceae: evidence from a complex multi-institutional KPC outbreak. PeerJ 6:e4210. doi: 10.7717/peerj.4210
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215(Suppl._1), S28–S36. doi: 10.1093/infdis/jiw282
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). Integrall: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nordmann, P., Poirel, L., and Dortet, L. (2012). Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18, 1503–1507. doi: 10.3201/eid1809.120355
Oelschlaeger, P., Mayo, S. L., and Pleiss, J. (2005). Impact of remote mutations on metallo-beta-lactamase substrate specificity: implications for the evolution of antibiotic resistance. Protein Sci. 14, 765–774. doi: 10.1110/ps.041093405
Ohno, Y., Nakamura, A., Hashimoto, E., Matsutani, H., Abe, N., Fukuda, S., et al. (2017). Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J. Infect. Chemother. 23, 224–229. doi: 10.1016/j.jiac.2016.12.013
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Poirel, L., Heritier, C., Tolun, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Ribeiro, V. B., Zavascki, A. P., Rozales, F. P., Pagano, M., Magagnin, C. M., Nodari, C. S., et al. (2014). Detection of bla(GES-5) in carbapenem-resistant kluyvera intermedia isolates recovered from the hospital environment. Antimicrob. Agents Chemother. 58, 622–623. doi: 10.1128/AAC.02271-13
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Segawa, T., Matsui, M., Suzuki, M., Tsutsui, A., Kuroda, M., Shibayama, K., et al. (2017). Utilizing the Carba NP test as an indicator of expression level of carbapenemase genes in Enterobacteriaceae. J. Microbiol. Methods 133, 35–39. doi: 10.1016/j.mimet.2016.12.015
Shibata, N., Doi, Y., Yamane, K., Yagi, T., Kurokawa, H., Shibayama, K., et al. (2003). PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 41, 5407–5413. doi: 10.1128/JCM.41.12.5407-5413.2003
Tanizawa, Y., Fujisawa, T., and Nakamura, Y. (2018). DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/bioinformatics/btx713
Tetz, G., Vecherkovskaya, M., Zappile, P., Dolgalev, I., Tsirigos, A., Heguy, A., et al. (2017). Complete genome sequence of Kluyvera intestini sp. nov., isolated from the stomach of a patient with gastric cancer. Genome Announc. 5:e01184-17. doi: 10.1128/genomeA.01184-17
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, L., Jing, Y., Lai, K., An, J., and Yang, J. (2018). A case of biliary tract infection caused by KPC-2-producing Kluyvera ascorbata. Case Rep. Infect. Dis. 2018:5745708. doi: 10.1155/2018/5745708
Woerther, P. L., Burdet, C., Chachaty, E., and Andremont, A. (2013). Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin. Microbiol. Rev. 26, 744–758. doi: 10.1128/CMR.00023-13
World Health Organization. (2017). Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities. Geneva: World Health Organization.
Yamamoto, N., Asada, R., Kawahara, R., Hagiya, H., Akeda, Y., Shanmugakani, R. K., et al. (2017). Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J. Hosp. Infect. 97, 212–217. doi: 10.1016/j.jhin.2017.07.015
Yoshino, Y., Nakazawa, S., Otani, S., Sekizuka, E., and Ota, Y. (2016). Nosocomial bacteremia due to Kluyvera cryocrescens: case report and literature review. ID Cases 4, 24–26. doi: 10.1016/j.idcr.2016.02.007
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Kluyvera, blaIMP-6, carbapenemase, IncN, Metakosakonia
Citation: Sekizuka T, Matsui M, Takahashi T, Hayashi M, Suzuki S, Tokaji A and Kuroda M (2018) Complete Genome Sequence of blaIMP–6-Positive Metakosakonia sp. MRY16-398 Isolate From the Ascites of a Diverticulitis Patient. Front. Microbiol. 9:2853. doi: 10.3389/fmicb.2018.02853
Received: 31 July 2018; Accepted: 06 November 2018;
Published: 22 November 2018.
Edited by:
Yi-Wei Tang, Memorial Sloan Kettering Cancer Center, United StatesReviewed by:
Ghassan M. Matar, American University of Beirut, LebanonFilipa Grosso, Universidade do Porto, Portugal
Copyright © 2018 Sekizuka, Matsui, Takahashi, Hayashi, Suzuki, Tokaji and Kuroda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Makoto Kuroda, makokuro@niid.go.jp
 Tsuyoshi Sekizuka
Tsuyoshi Sekizuka Mari Matsui2
Mari Matsui2 Satowa Suzuki
Satowa Suzuki Makoto Kuroda
Makoto Kuroda