- State Key Laboratory of Genetic Engineering, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, China
At high vegetation density, shade-intolerant plants sense a reduction in the red (660 nm) to far-red (730 nm) light ratio (R/FR) in addition to a general reduction in light intensity. These light signals trigger a spectrum of morphological changes manifested by growth of stem-like tissue (hypocotyl, petiole, etc.) instead of harvestable organs (leaves, fruits, seeds, etc.)—namely, shade avoidance syndrome (SAS). Common phenotypical changes related to SAS are changes in leaf hyponasty, an increase in hypocotyl and internode elongation and extended petioles. Prolonged shade exposure leads to early flowering, less branching, increased susceptibility to insect herbivory, and decreased seed yield. Thus, shade avoidance significantly impacts on agronomic traits. Many genetic and molecular studies have revealed that phytochromes, cryptochromes and UVR8 (UV-B photoreceptor protein) monitor the changes in light intensity under shade and regulate the stability or activity of phytochrome-interacting factors (PIFs). PIF-governed modulation of the expression of auxin biosynthesis, transporter and signaling genes is the major driver for shade-induced hypocotyl elongation. Besides auxin, gibberellins, brassinosteroids, and ethylene are also required for shade-induced hypocotyl or petiole elongation growth. In leaves, accumulated auxin stimulates cytokinin oxidase expression to break down cytokinins and inhibit leaf growth. In the young buds, shade light promotes the accumulation of abscisic acid to repress branching. Shade light also represses jasmonate- and salicylic acid-induced defense responses to balance resource allocation between growth and defense. Here we will summarize recent findings relating to such hormonal regulation in SAS in Arabidopsis thaliana, Brassica rapa, and certain crops.
Introduction
Over the past few decades, a substantial body of studies has focused on understanding how plants sense the proximity of neighbors, how they respond at molecular levels, and how they adjust their morphological and physiological indexes. Many important light signaling components have been shown to regulate the shade avoidance responses—for example, PIFs (phytochrome interacting factors), HFR1 (long hypocotyl in far-red 1), PAR1/2 (phytochrome rapidly regulated 1/2) and COP1 (constitutive photomorphogenic 1). Meanwhile, various phytohormones are also involved and coordinated to shape shade-regulated plant architecture. Analyses of hormonal biosynthetic and signaling mutants, combined with studies of exogenous hormone applications, have implicated the roles of these phytohormones in multiple shade avoidance responses. In this review, we provide an overview of the current understanding of shade light and subsequent hormonal regulation.
Shade Signal and Plant Perception
Light-quality signals are of paramount importance in detecting neighboring vegetation. Photosynthetic pigments in leaves absorb strongly in the range of photosynthetically active radiation (PAR) (400–700 nm) and UV radiation (280–400 nm), and reflect far-red wavelength (700–800 nm) (Casal, 2013). Thus, natural shade is a combination of the reduction in the red/far-red ratio (R/FR), the reduction in red plus far-red irradiance, the reduction in blue and UV irradiance, and the reduced blue/green ratio. To detect these spectral differences, plants use multiple light sensors, such as red and far-red light absorbing phytochromes, the blue/UV-A light sensing cryptochromes, and the UV-B photoreceptor protein (UVR8).
A Brief Account of the Shade Signaling Pathway
Phytochromes exist in two photoconvertible forms: an inactive R-absorbing Pr form and an active FR-absorbing Pfr form. The steady-state ratio of Pr and Pfr forms depends on R/FR. The constitutive shade avoidance syndrome (SAS) phenotype of Arabidopsis phyB mutant plants indicates that phyB plays a dominant role in inhibiting SAS (Franklin and Quail, 2010). High R/FR establishes a high proportion of phyB Pfr, which interacts with the bHLH family of transcription factor PIFs and triggers the phosphorylation, ubiquitination and degradation of PIFs. In contrast, low R/FR drives Pfr-to-Pr conversion and releases the suppression of PIFs. Activated PIFs promote gene expression related to shade-induced growth. PIF7, PIF4 and PIF5 play central roles in this process (Lorrain et al., 2008; Li L. et al., 2012).
To prevent exaggerated shade-avoidance responses, shade-induced HFR1 (Sessa et al., 2005; Hornitschek et al., 2009), PAR1/2 (Roig-Villanova et al., 2006; Galstyan et al., 2011; Bou-Torrent et al., 2014), and PIL1 (PIF3 like 1) (Li et al., 2014; Luo et al., 2014) are proposed as the negative regulators of PIFs. The bZIP transcription factor, elongated hypocotyl 5 (HY5), is also reported to form non-functional complexes with PIFs (Chen et al., 2013; Toledo-Ortiz et al., 2014). In addition to directly binding with PIFs, the Suppressor of phyA-105 (SPA)/COP1 E3 ubiquitin ligase complex indirectly enhances PIF activity by degrading HFR1 and HY5 to augment shade responses (Sheerin et al., 2015; Pacin et al., 2016). BBX (double B-box) 21 and BBX25 regulate shade response through the function in the COP1 signaling pathway (Crocco et al., 2010; Gangappa et al., 2013).
Cryptochromes (CRYs) are involved in repressing a low blue-mediated SAS by regulating PIF abundance and activity (de Wit et al., 2016; Pedmale et al., 2016). PIF activity is enhanced directly through CRY inactivation and indirectly through relieved inhibition of COP1, which increases the degradation of negative regulators of PIF, including HFR1 and HY5 (de Wit et al., 2016).
UV-B-mediated inhibition of shade responses has been reported to occur through the degradation of PIF4/5 (Hayes et al., 2014).
In summary, downstream of photoreceptors, PIFs, as the key regulators, determine the massive transcriptional reprogramming upon perception of shade light, and also mediates the convergence between light and hormones.
Auxin, A Prominent Player in Shade-Induced Elongation Growth
A forward genetic screen for impaired shade-induced hypocotyl elongation in Arabidopsis identified TAA1, an enzyme catalyzing the first step of an auxin biosynthetic pathway (Tao et al., 2008; Won et al., 2011). Later, a family of enzymes encoded by YUCCA (YUC) genes has been functionally positioned as the second and rate-limiting step of TAA1-dependent auxin biosynthesis (the indole-2-pyruvic acid pathway, or “IPA pathway”). The transcriptional regulation of YUCCA genes by PIF7 has been found to link photoperception with auxin biosynthesis (Li L. et al., 2012). The level of shade-stimulated free indole-3-acetic acid (IAA) is blunted in taa1, and pif7 mutants confirm that auxin production through the TAA1-YUC pathway is required to initiate the SAS in seedlings (Tao et al., 2008; Li L. et al., 2012; Procko et al., 2014). PIF4 and PIF5 are partially redundant, with PIF7 regulating the expression of YUCCA genes (Hornitschek et al., 2012). Correspondingly, the yuc2 yuc5 yuc8 yuc9 quadruple mutant displays the completely disrupted SAS (Nozue et al., 2015; Muller-Moule et al., 2016). Tissue-level measurement in Brassica rapa seedlings has suggested that auxin appears to be generated in the cotyledons and transported to the hypocotyl (Procko et al., 2014). Indeed, seedlings treated with the auxin transport inhibitor naphthylphalamic acid (NPA) totally abolish shade-induced hypocotyl elongation (Tao et al., 2008). Consistently, pin3-3 (PIN3, auxin transporter) exhibits an impaired shade-induced hypocotyl elongation (Keuskamp et al., 2010), and the mutation in SAV4 leads to defective basipetal auxin transport and shade responses (Ge et al., 2017), indicating that auxin redistribution is important for shade-avoidance reactions (Morelli and Ruberti, 2000).
Besides auxin biosynthesis and transport, auxin sensitivity is also enhanced under shade (Nozue et al., 2011; Hornitschek et al., 2012; Bou-Torrent et al., 2014). Auxin signaling components, such as AUX/IAAs (Auxin/indole-3-acetic acid), have been reported to modulate the SAS (Steindler et al., 1999; Procko et al., 2016).
In addition to Arabidopsis, the key role of auxin on the SAS has also been confirmed in crop species (Carriedo et al., 2016). Shade-induced changes in auxin level have been found in sunflower (Kurepin et al., 2007) and tomato (Kozuka et al., 2010). Expression quantitative trait locus (eQTL) analysis identified a group of auxin-related genes, which were down-regulated in shade-tolerant tomato lines and up-regulated in the shade responders, suggesting the role of auxin in the natural variation of the SAS (Bush et al., unpublished). In maize seedlings (Wang et al., 2016) and rice seedlings (Liu et al., 2016), the expression of auxin-responsive genes is also dramatically affected by shade treatment.
Considered together, it may be concluded that intact auxin biosynthesis, transportation and signaling are required for shade-induced stem growth.
Gibberellin, Another Shade Growth-Promoting Hormone
Shade treatment resulted in an increased gibberellin (GA) concentration in bean internode (Beall et al., 1996), cowpea (Vigna sinensis) epicotyls (Martínez-García et al., 2000), sunflower stem (Kurepin et al., 2007) and Arabidopsis seedling (Bou-Torrent et al., 2014). The shade-induced GA biosynthetic enzymes GA20ox1, GA20ox2, and GA3ox at least in part account for the increase in active GA (Hisamatsu et al., 2005; Yu et al., 2015).
Bioactive GA leads to proteasomal degradation of DELLA proteins (Harberd et al., 2009). Lacking direct DNA binding capability, DELLAs are direct interactors of PIFs. Their binding prevents PIF proteins from binding DNA and thus negatively regulates the expression of genes involved in cell elongation (de Lucas et al., 2008; Feng et al., 2008). Shade-induced breakdown of DELLA proteins due to increased gibberellin biosynthesis releases the suppression of PIFs, and activates the transcription of target genes. The GA-insensitive gai gain-of-function mutant, which has a stable GAI (DELLA) protein, shows a reduced SAS (Djakovic-Petrovic et al., 2007), suggesting that DELLA proteins constrain the SAS.
It is noteworthy that proteins that physically interact with DELLA proteins may alleviate DELLA-mediated repression of PIF activity, such as BBX24. The shade-response defect in bbx24 mutants is rescued by a GA treatment (Crocco et al., 2015).
In addition to GA-induced seedling phenotypes, GA biosynthesis and signaling are also important for shade-induced flowering. Silencing GA20ox2 expression delays flowering of Arabidopsis exposed to a FR-enriched light condition (Hisamatsu and King, 2008).
Ethylene, An Organ-Specific Regulator of the SAS
Low R/FR can enhance the production of ethylene in wide-type tobacco (Pierik et al., 2004). In Arabidopsis, shade-induced petiole elongation was absent in the ethylene-insensitive mutants ein2-1 and ein3-1eil1-3, indicating that ethylene is a positive regulator of shade-induced petiole elongation (Pierik et al., 2009). However, the ein3eil1 mutant retains a full shade-induced hypocotyl response (Das et al., 2016). The controversy suggests that ethylene plays a role in organ-specific shade response.
A recent research shows that light activation of photoreceptor phyB results in rapid degradation of EIN3, a master transcription factor in the ethylene signaling pathway (Shi et al., 2016). The position of ethylene signaling components under shade is worthy of further investigation.
Brassinosteroid, A Dynamic Regulator Under Shade
The promotion of stem growth by shade light requires brassinosteroids (BRs) because the BR biosynthesis mutant dwarf1 (Luccioni et al., 2002) and rot3 (Kim et al., 1998) are unable to show the elongation of hypocotyl under shade, as with wild-type seedlings treated with the BR synthesis inhibitor brassinazole (Keuskamp et al., 2011). BR biosynthesis is also required for petiole growth under low R/FR (Kozuka et al., 2010). However, short-term (4 h) simulated shade treatments resulted in lower levels of the active BR, and longer periods (24 h) abolished the differences in BR levels in whole seedlings (Bou-Torrent et al., 2014), suggesting that simulated shade altered BR levels in a dynamic fashion.
Beside the level of hormones, the sensitivity of seedlings to hormones also has an important effect on shade-induced growth. BR signaling components BR-ENHANCED EXPRESSION (BEE) and BES1-INTERACTING MYC-LIKE (BIM) are positive regulators of SAS hypocotyl responses because bee123 and bim123 seedlings display hypocotyl elongation defects after detecting simulated shade (Cifuentes-Esquivel et al., 2013). Remarkably, DELLAs negatively regulate BR signaling by binding BZR1 and reducing the expression of BR-responsive genes (Bai et al., 2012; Gallego-Bartolome et al., 2012; Li Q.F. et al., 2012). The transcription factor BZR1 and PIF4 physically interact and synergistically regulate target genes (Oh et al., 2012; Kohnen et al., 2016). Given that the binding of DELLA and PIFs impair the DNA-binding ability of PIFs, the complex of DELLAs, BZR1, and PIFs may play a role in stem elongation, and possibly exerts a similar function in shade avoidance, but this needs further investigation (Casal, 2013; de Lucas and Prat, 2014). In concordance with these findings, BR-responsive genes are overrepresented in end-of-day FR-induced genes in both the leaf blade and petiole (Kozuka et al., 2010). Although the majority of the BR genomic response comprises genes annotated as auxin responsive, the regulation of BR and auxin on SAS responses might nevertheless occur in a non-redundant and non-synergistic manner, because the response to blue light depletion will be fully inhibited only when both hormones are blocked simultaneously (Keuskamp et al., 2011).
In particular, the BR response appears to be required for the full expression of the SAS phenotypes under low blue light (Keller et al., 2011; Keuskamp et al., 2011). The question as to how BR biosynthesis and signaling dynamically respond to low R/FR or low blue light is yet to be answered.
Cytokinin, Ensuring Reallocation of Plant Resources
The role of cytokinins (CKs) in shade avoidance responses was discovered from the response of plants to vertical light intensity gradients in leaf canopies (Pons et al., 2001). In shaded leaves, where stomatal conductance and transpiration rate are reduced, the low delivery rate of CKs leads to reduced photosynthetic capacity and ultimately senescence (Boonman and Pons, 2007).
Another role of CKs was found in the inhibition of leaf growth in shade. Low R/FR signal can induce hypocotyl elongation and also trigger a rapid arrest of leaf-primordia growth by the breakdown of auxin-induced CKs through the action of AtCKX6 (cytokinin oxidase) in the incipient vein cells of developing primordia (Carabelli et al., 2007). In addition, the CK receptor AHK3 has been reported to mediate the root-to-hypocotyl ratio response under shade conditions (Novak et al., 2015).
The reduction of bioactive CKs triggers a reduced photosynthetic capacity and a transient arrest of leaf development, ensuring that energy resources are indeed redirected into extension growth in shade.
Jasmonic Acid, Shade-Reduced Hormone Related to Defense
Plants often display a weak defense in insect and pathogen infection under shade conditions or FR-enriched conditions (Cerrudo et al., 2012; de Wit et al., 2013; Ballare, 2014). Shade has been shown to reduce herbivory-induced jasmonic acid (JA) accumulation (Agrawal et al., 2012), and FR-exposed plants suffer more insect herbivory than wild-type plants (Moreno et al., 2009), suggesting that shade can down-regulate the JA pathway to control plant immunity.
The JAZ-DELLA pathway is an important modulator of plant immunity under shade conditions (Moreno and Ballare, 2014). DELLA proteins positively regulate JA signaling by interacting with JAZs, and this interaction weakens the ability of JAZs to repress MYC2 (Hou et al., 2010; Yang et al., 2012). As described previously, DELLA proteins negatively regulate growth-related genes by binding PIFs (de Lucas et al., 2008; Feng et al., 2008). JAZ10 is required for the inhibitory effect of shade on JA responses (Leone et al., 2014). Therefore, shade conditions induce GA synthesis and the degradation of DELLA proteins, consequently increasing PIF-dependent growth and impairing JAZ-dependent defense. Canopy shade represses JA-mediated defenses via shade-induced stabilization of JAZ proteins and triggers inactivation of MYC2, MYC3, and MYC4 proteins (Chico et al., 2014). By contrast, regulation of the protein stability of MYCs and JAZs by shade facilitates reallocation of resources from defense to growth. The mutants deficient in JA biosynthesis and signaling display exaggerated shade-induced hypocotyl responses to a low R/FR ratio (Robson et al., 2010). Moreover, several FR light induced gene expressions are dependent on CORONATINE INSENSITIVE1 (COI1), a central component of JA signaling (Robson et al., 2010).
Canopy light cues affect emission of constitutive and methyl JA-induced volatile organic compounds, which can be detected by herbivorous insects (Kegge et al., 2013). A recent study found that in tomato (Solanum lycopersicum) phyB inactivation led the plants to produce a blend of JA-induced monoterpenes that increased their attractiveness to the predatory mirid bug Macrolophus pygmaeus (Cortes et al., 2016; Ballare and Pierik, 2017).
Certain transcription factors in the JA signaling pathway also participate in the regulation of SAS; for example, PHYTOCHROME AND FLOWERING TIME 1 (PFT1), a subunit of Mediator, is required for both JA-dependent defense gene expression and shade-induced early flowering (Cerdan and Chory, 2003; Cevik et al., 2012; Inigo et al., 2012). These factors could be the additional linkers of light signal and JA-mediated defenses.
Salicylic Acid, Another Shade-Reduced Hormone
Salicylic acid (SA)-dependent disease resistance is also reduced under shade, which is considered as the early warning signal for plant competition (de Wit et al., 2013). Reduced SA synthesis (Griebel and Zeier, 2008) and response (de Wit et al., 2013) have been correlated with phyB inactivation. Under a low R/FR ratio, the phosphorylation level of the SA-signaling component NONEXPRESSOR of PATHOGENESIS-RELATED GENE 1 (NPR1) is reduced, which partly explains why shade reduces SA-dependent disease resistance. A more detailed explanation of the mechanism that exists between shade avoidance responses and SA is required.
Abscisic Acid, Repressing Branching Under Shade
Abscisic acid (ABA) is commonly known as the “stress hormone” that responds to a variety of environmental stresses including both biotic and abiotic stress. Shade conditions increase ABA levels in sunflower (Helianthus annuus) (Kurepin et al., 2007) and tomato leaves (Cagnola et al., 2012). Shade increases the endogenous ABA level probably by enhancing the transcript levels of ABA biosynthetic gene NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3) and NCED5, particularly in hypocotyls (Kohnen et al., 2016). Several ABA signaling genes (ABF3, AFP1, AFP3, and GBF3) are up-regulated by a neighbor signal (Sellaro et al., 2017).
Shade light exerts a strong influence on branch development (Finlayson et al., 2010; Su et al., 2011). One recent study suggested that shade represses branching in bud n-2 by accumulation of ABA (Reddy et al., 2013). The genes involved in ABA biosynthesis and signal transduction showed varied gene expression patterns in responsive buds with increasing R/FR treatment. ABA biosynthesis mutants (nced3-2 and aba2-1) exhibited enhanced branching capacity under low R/FR.
However, ABA was not involved in shade-induced petiole elongation (Pierik et al., 2011), suggesting that the roles of ABA in the SAS may be organ specific.
Strigolactone, An Unclear Role in the SAS
Most shade-avoiding plants display reduced branching and enhanced apical growth, which helps them to compete for incident light. Strigolactone (SL) is one of the hormones that control lateral shoot growth. In Arabidopsis, BRC1 (BRANCHED1) is up-regulated in the axillary buds of plants grown at high density and is required for shade-mediated branch suppression (Aguilar-Martinez et al., 2007; Gonzalez-Grandio et al., 2013). In sorghum, inhibition of outgrowth in a phyB mutant and by FR treatment is correlated with an increase in the transcript levels of the SL-signaling gene SbMAX2 in buds (Kebrom et al., 2010). The involvement of SL in SAS has been observed, but more detailed studies of this mechanism are required.
Besides branching, Arabidopsis max2 mutants show longer hypocotyls under red, far-red and blue light than wild–type plants (Shen et al., 2012; Jia et al., 2014). The double mutant pif1max2 shows a similar hypocotyl length to max2, which indicates that MAX2 is epistatic to PIF1 (Shen et al., 2012). MAX2 plays a role in the light signaling pathway, but further investigation of the mechanisms involved is needed.
Karrikins, A Possible Way to Attenuate the SAS
Studies have shown that karrikins enhance the sensitivity of seedlings to light (Waters and Smith, 2013). Since karrikins can inhibit elongation of the hypocotyl and increase the chlorophyll content (Nelson et al., 2010), they may be an efficient solution to attenuating plant SAS during the seedling stage (Meng et al., 2016).
Final Remarks
This review focused on understanding the interaction between phytohormones and the SAS (Figure 1). The regulations of these phytohormones on the SAS described here might vary according to tissue type (Kohnen et al., 2016), stage of development (Roig-Villanova and Martinez-Garcia, 2016) and species (Liu et al., 2016). In this regard, further research into the spatial and temporal regulation of phytohormones is necessary for a mechanistical understanding of the SAS. Moreover, crosstalk among hormones under shade conditions is also worthy of further investigation.
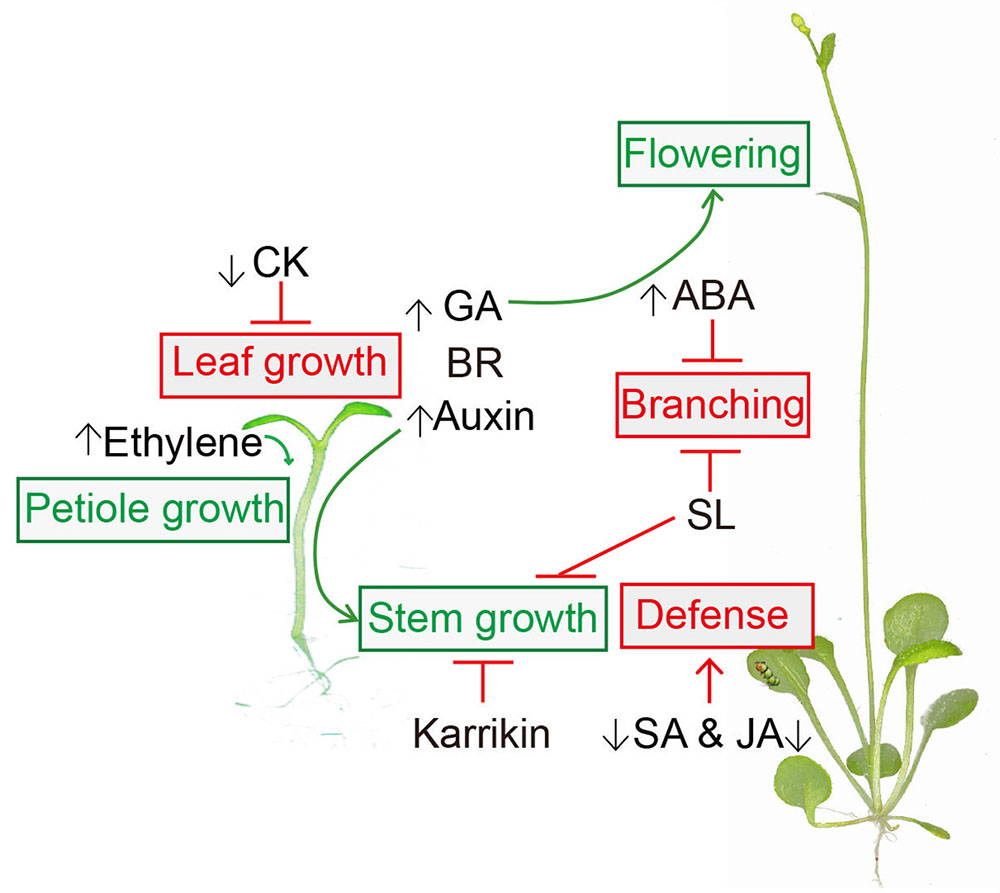
FIGURE 1. Hormonal regulation in shade avoidance. Auxin, Gibberellin (GA), Brassinosteroid (BR), Karrikin and strigolactone (SL) are involved in shade-regulated stem growth. Ethylene is required for shade-induced petiole elongation. Shade-reduced cytokinin (CK) inhibits the leaf growth. Shade light also represses salicylic acid (SA) and jasmonic acid (JA) mediated defense. Abscisic acid (ABA) and SL suppress branching in shade. GA contributes to shade-induced early flowering. Shade-stimulations are presented in green and shade-supressions are presented in red.
Author Contributions
CY and LL designed and wrote the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China SQ2017YFJC040047-01 and the National Natural Science Foundation of China Grants 31470374 and 31500973.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very thankful to Christian Fankhauser (University of Lausanne, Switzerland) for his critical comments on the manuscript.
References
Agrawal, A. A., Kearney, E. E., Hastings, A. P., and Ramsey, T. E. (2012). Attenuation of the jasmonate burst, plant defensive traits, and resistance to specialist monarch caterpillars on shaded common milkweed (Asclepias syriaca). J. Chem. Ecol. 38, 893–901. doi: 10.1007/s10886-012-0145-143
Aguilar-Martinez, J. A., Poza-Carrion, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Bai, M. Y., Shang, J. X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Ballare, C. L. (2014). Light regulation of plant defense. Annu. Rev. Plant Biol. 65, 335–363. doi: 10.1146/annurev-arplant-050213-40145
Ballare, C. L., and Pierik, R. (2017). The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. doi: 10.1111/pce.12914 [Epub ahead of print].
Beall, F. D., Yeung, E. C., and Pharis, R. P. (1996). Far-red light stimulates internode elongation, cell division, cell elongation, and gibberellin levels in bean. Can. J. Bot. 74, 743–752. doi: 10.1139/b96-093
Boonman, A., and Pons, T. L. (2007). Canopy light gradient perception by cytokinin. Plant Signal. Behav. 2, 489–491.
Bou-Torrent, J., Galstyan, A., Gallemi, M., Cifuentes-Esquivel, N., Molina-Contreras, M. J., Salla-Martret, M., et al. (2014). Plant proximity perception dynamically modulates hormone levels and sensitivity in Arabidopsis. J. Exp. Bot. 65, 2937–2947. doi: 10.1093/jxb/eru083
Cagnola, J. I., Ploschuk, E., Benech-Arnold, T., Finlayson, S. A., and Casal, J. J. (2012). Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol. 160, 1110–1119. doi: 10.1104/pp.112.201921
Carabelli, M., Possenti, M., Sessa, G., Ciolfi, A., Sassi, M., Morelli, G., et al. (2007). Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 21, 1863–1868. doi: 10.1101/gad.432607
Carriedo, L. G., Maloof, J. N., and Brady, S. M. (2016). Molecular control of crop shade avoidance. Curr. Opin. Plant Biol. 30, 151–158. doi: 10.1016/j.pbi.2016.03.005
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221.
Cerdan, P. D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. doi: 10.1038/nature01636
Cerrudo, I., Keller, M. M., Cargnel, M. D., Demkura, P. V., de Wit, M., Patitucci, M. S., et al. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158, 2042–2052. doi: 10.1104/pp.112.193359
Cevik, V., Kidd, B. N., Zhang, P., Hill, C., Kiddle, S., Denby, K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. doi: 10.1104/pp.112.202697
Chen, D., Xu, G., Tang, W., Jing, Y., Ji, Q., Fei, Z., et al. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25, 1657–1673. doi: 10.1105/tpc.112.104869
Chico, J. M., Fernandez-Barbero, G., Chini, A., Fernandez-Calvo, P., Diez-Diaz, M., and Solano, R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26, 1967–1980. doi: 10.1105/tpc.114.125047
Cifuentes-Esquivel, N., Bou-Torrent, J., Galstyan, A., Gallemi, M., Sessa, G., Salla Martret, M., et al. (2013). The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 75, 989–1002. doi: 10.1111/tpj.12264
Cortes, L. E., Weldegergis, B. T., Boccalandro, H. E., Dicke, M., and Ballare, C. L. (2016). Trading direct for indirect defense? Phytochrome B inactivation in tomato attenuates direct anti-herbivore defenses whilst enhancing volatile-mediated attraction of predators. New Phytol. 212, 1057–1071. doi: 10.1111/nph.14210
Crocco, C. D., Holm, M., Yanovsky, M. J., and Botto, J. F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64, 551–562. doi: 10.1111/j.1365-313X.2010.04360.x
Crocco, C. D., Locascio, A., Escudero, C. M., Alabadi, D., Blazquez, M. A., and Botto, J. F. (2015). The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat. Commun. 6, 6202. doi: 10.1038/ncomms7202
Das, D., St Onge, K. R., Voesenek, L. A., Pierik, R., and Sasidharan, R. (2016). Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol. 172, 718–733. doi: 10.1104/pp.16.00725
de Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
de Lucas, M., and Prat, S. (2014). PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 202, 1126–1141. doi: 10.1111/nph.12725
de Wit, M., Keuskamp, D. H., Bongers, F. J., Hornitschek, P., Gommers, C. M., Reinen, E., et al. (2016). Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 26, 3320–3326. doi: 10.1016/j.cub.2016.10.031
de Wit, M., Spoel, S. H., Sanchez-Perez, G. F., Gommers, C. M., Pieterse, C. M., Voesenek, L. A., et al. (2013). Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 75, 90–103. doi: 10.1111/tpj.12203
Djakovic-Petrovic, T., de Wit, M., Voesenek, L. A., and Pierik, R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51, 117–126. doi: 10.1111/j.1365-313X.2007.03122.x
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Finlayson, S. A., Krishnareddy, S. R., Kebrom, T. H., and Casal, J. J. (2010). Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 152, 1914–1927. doi: 10.1104/pp.109.148833
Franklin, K. A., and Quail, P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. doi: 10.1093/jxb/erp304
Gallego-Bartolome, J., Minguet, E. G., Grau-Enguix, F., Abbas, M., Locascio, A., Thomas, S. G., et al. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 13446–13451. doi: 10.1073/pnas.1119992109
Galstyan, A., Cifuentes-Esquivel, N., Bou-Torrent, J., and Martinez-Garcia, J. F. (2011). The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 66, 258–267. doi: 10.1111/j.1365-313X.2011.04485.x
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Ge, Y., Yan, F., Zourelidou, M., Wang, M., Ljung, K., Fastner, A., et al. (2017). SHADE AVOIDANCE 4 is required for proper auxin distribution in the hypocotyl. Plant Physiol. 173, 788–800. doi: 10.1104/pp.16.01491
Gonzalez-Grandio, E., Poza-Carrion, C., Sorzano, C. O., and Cubas, P. (2013). BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25, 834–850. doi: 10.1105/tpc.112.108480
Griebel, T., and Zeier, J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147, 790–801. doi: 10.1104/pp.108.119503
Harberd, N. P., Belfield, E., and Yasumura, Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21, 1328–1339. doi: 10.1105/tpc.109.066969
Hayes, S., Velanis, C. N., Jenkins, G. I., and Franklin, K. A. (2014). UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. U.S.A. 111, 11894–11899. doi: 10.1073/pnas.1403052111
Hisamatsu, T., and King, R. W. (2008). The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot. 59, 3821–3829. doi: 10.1093/jxb/ern232
Hisamatsu, T., King, R. W., Helliwell, C. A., and Koshioka, M. (2005). The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 138, 1106–1116. doi: 10.1104/pp.104.059055
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., Lopez-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Hornitschek, P., Lorrain, S., Zoete, V., Michielin, O., and Fankhauser, C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. doi: 10.1038/emboj.2009.306
Hou, X., Lee, L. Y., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894. doi: 10.1016/j.devcel.2010.10.024
Inigo, S., Alvarez, M. J., Strasser, B., Califano, A., and Cerdan, P. D. (2012). PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 69, 601–612. doi: 10.1111/j.1365-313X.2011.04815.x
Jia, K. P., Luo, Q., He, S. B., Lu, X. D., and Yang, H. Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant 7, 528–540. doi: 10.1093/mp/sst093
Kebrom, T. H., Brutnell, T. P., and Finlayson, S. A. (2010). Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33, 48–58. doi: 10.1111/j.1365-3040.2009.02050.x
Kegge, W., Weldegergis, B. T., Soler, R., Vergeer-Van Eijk, M., Dicke, M., Voesenek, L. A., et al. (2013). Canopy light cues affect emission of constitutive and methyl jasmonate-induced volatile organic compounds in Arabidopsis thaliana. New Phytol. 200, 861–874. doi: 10.1111/nph.12407
Keller, M. M., Jaillais, Y., Pedmale, U. V., Moreno, J. E., Chory, J., and Ballare, C. L. (2011). Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67, 195–207. doi: 10.1111/j.1365-313X.2011.04598.x
Keuskamp, D. H., Pollmann, S., Voesenek, L. A., Peeters, A. J., and Pierik, R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. U.S.A. 107, 22740–22744. doi: 10.1073/pnas.1013457108
Keuskamp, D. H., Sasidharan, R., Vos, I., Peeters, A. J., Voesenek, L. A., and Pierik, R. (2011). Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 67, 208–217. doi: 10.1111/j.1365-313X.2011.04597.x
Kim, G. T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391.
Kohnen, M. V., Schmid-Siegert, E., Trevisan, M., Petrolati, L. A., Senechal, F., Muller-Moule, P., et al. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28, 2889–2904. doi: 10.1105/tpc.16.00463
Kozuka, T., Kobayashi, J., Horiguchi, G., Demura, T., Sakakibara, H., Tsukaya, H., et al. (2010). Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 153, 1608–1618. doi: 10.1104/pp.110.156802
Kurepin, L. V., Emery, R. J., Pharis, R. P., and Reid, D. M. (2007). Uncoupling light quality from light irradiance effects in Helianthus annuus shoots: putative roles for plant hormones in leaf and internode growth. J. Exp. Bot. 58, 2145–2157. doi: 10.1093/jxb/erm068
Leone, M., Keller, M. M., Cerrudo, I., and Ballare, C. L. (2014). To grow or defend? Low red : far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 204, 355–367. doi: 10.1111/nph.12971
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Li, L., Zhang, Q., Pedmale, U. V., Nito, K., Fu, W., Lin, L., et al. (2014). PIL1 participates in a negative feedback loop that regulates its own gene expression in response to shade. Mol. Plant 7, 1582–1585. doi: 10.1093/mp/ssu068
Li, Q. F., Wang, C., Jiang, L., Li, S., Sun, S. S., and He, J. X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5, ra72. doi: 10.1126/scisignal.2002908
Liu, H., Yang, C., and Li, L. (2016). Shade-induced stem elongation in rice seedlings: implication of tissue-specific phytohormone regulation. J. Integr. Plant Biol. 58, 614–617. doi: 10.1111/jipb.12468
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Luccioni, L. G., Oliverio, K. A., Yanovsky, M. J., Boccalandro, H. E., and Casal, J. J. (2002). Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 128, 173–181.
Luo, Q., Lian, H. L., He, S. B., Li, L., Jia, K. P., and Yang, H. Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441–2456. doi: 10.1105/tpc.113.121657
Martínez-García, J. F., Santes, C. M., and García-Martínez, J. L. (2000). The end-of-day far-red irradiation increases gibberellin A1 content in cowpea (Vigna sinensis) epicotyls by reducing its inactivation. Physiol. Plant. 108, 426–434. doi: 10.1034/j.1399-3054.2000.t01-1-100413.x
Meng, Y., Shuai, H., Luo, X., Chen, F., Zhou, W., Yang, W., et al. (2016). Karrikins: regulators involved in phytohormone signaling networks during seed germination and seedling development. Front. Plant Sci. 7:2021. doi: 10.3389/fpls.2016.02021
Morelli, G., and Ruberti, I. (2000). Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol. 122, 621–626.
Moreno, J. E., and Ballare, C. L. (2014). Phytochrome regulation of plant immunity in vegetation canopies. J. Chem. Ecol. 40, 848–857. doi: 10.1007/s10886-014-0471-478
Moreno, J. E., Tao, Y., Chory, J., and Ballare, C. L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. U.S.A. 106, 4935–4940. doi: 10.1073/pnas.0900701106
Muller-Moule, P., Nozue, K., Pytlak, M. L., Palmer, C. M., Covington, M. F., Wallace, A. D., et al. (2016). YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4:e2574. doi: 10.7717/peerj.2574
Nelson, D. C., Flematti, G. R., Riseborough, J. A., Ghisalberti, E. L., Dixon, K. W., and Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 7095–7100. doi: 10.1073/pnas.0911635107
Novak, J., Cerny, M., Pavlu, J., Zemankova, J., Skalak, J., Plackova, L., et al. (2015). Roles of proteome dynamics and cytokinin signaling in root to hypocotyl ratio changes induced by shading roots of Arabidopsis seedlings. Plant Cell Physiol. 56, 1006–1018. doi: 10.1093/pcp/pcv026
Nozue, K., Harmer, S. L., and Maloof, J. N. (2011). Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 156, 357–372. doi: 10.1104/pp.111.172684
Nozue, K., Tat, A. V., Kumar Devisetty, U., Robinson, M., Mumbach, M. R., Ichihashi, Y., et al. (2015). Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genet. 11:e1004953. doi: 10.1371/journal.pgen.1004953
Oh, E., Zhu, J. Y., and Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. doi: 10.1038/ncb2545
Pacin, M., Semmoloni, M., Legris, M., Finlayson, S. A., and Casal, J. J. (2016). Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 211, 967–979. doi: 10.1111/nph.13965
Pedmale, U. V., Huang, S. S., Zander, M., Cole, B. J., Hetzel, J., Ljung, K., et al. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. doi: 10.1016/j.cell.2015.12.018
Pierik, R., Cuppens, M. L., Voesenek, L. A., and Visser, E. J. (2004). Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol. 136, 2928–2936. doi: 10.1104/pp.104.045120
Pierik, R., De Wit, M., and Voesenek, L. A. (2011). Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol. 189, 122–134. doi: 10.1111/j.1469-8137.2010.03458.x
Pierik, R., Djakovic-Petrovic, T., Keuskamp, D. H., de Wit, M., and Voesenek, L. A. (2009). Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol. 149, 1701–1712. doi: 10.1104/pp.108.133496
Pons, T. L., Jordi, W., and Kuiper, D. (2001). Acclimation of plants to light gradients in leaf canopies: evidence for a possible role for cytokinins transported in the transpiration stream. J. Exp. Bot. 52, 1563–1574.
Procko, C., Burko, Y., Jaillais, Y., Ljung, K., Long, J. A., and Chory, J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30, 1529–1541. doi: 10.1101/gad.283234.116
Procko, C., Crenshaw, C. M., Ljung, K., Noel, J. P., and Chory, J. (2014). Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 165, 1285–1301. doi: 10.1104/pp.114.241844
Reddy, S. K., Holalu, S. V., Casal, J. J., and Finlayson, S. A. (2013). Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 163, 1047–1058. doi: 10.1104/pp.113.221895
Robson, F., Okamoto, H., Patrick, E., Harris, S. R., Wasternack, C., Brearley, C., et al. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160. doi: 10.1105/tpc.109.067728
Roig-Villanova, I., Bou, J., Sorin, C., Devlin, P. F., and Martinez-Garcia, J. F. (2006). Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141, 85–96. doi: 10.1104/pp.105.076331
Roig-Villanova, I., and Martinez-Garcia, J. F. (2016). Plant responses to vegetation proximity: a whole life avoiding shade. Front. Plant Sci. 7:236. doi: 10.3389/fpls.2016.00236
Sellaro, R., Pacin, M., and Casal, J. J. (2017). Meta-analysis of the transcriptome reveals a core set of shade-avoidance genes in Arabidopsis. Photochem. Photobiol. 93, 692–702. doi: 10.1111/php.12729
Sessa, G., Carabelli, M., Sassi, M., Ciolfi, A., Possenti, M., Mittempergher, F., et al. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19, 2811–2815. doi: 10.1101/gad.364005
Sheerin, D. J., Menon, C., zur Oven-Krockhaus, S., Enderle, B., Zhu, L., Johnen, P., et al. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189-201. doi: 10.1105/tpc.114.134775.
Shen, H., Zhu, L., Bu, Q. Y., and Huq, E. (2012). MAX2 affects multiple hormones to promote photomorphogenesis. Mol. Plant 5, 750–762. doi: 10.1093/mp/sss029
Shi, H., Shen, X., Liu, R., Xue, C., Wei, N., Deng, X. W., et al. (2016). The Red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev. Cell 39, 597–610. doi: 10.1016/j.devcel.2016.10.020
Steindler, C., Matteucci, A., Sessa, G., Weimar, T., Ohgishi, M., Aoyama, T., et al. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126, 4235–4245.
Su, H., Abernathy, S. D., White, R. H., and Finlayson, S. A. (2011). Photosynthetic photon flux density and phytochrome B interact to regulate branching in Arabidopsis. Plant Cell Environ. 34, 1986–1998. doi: 10.1111/j.1365-3040.2011.02393.x
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Toledo-Ortiz, G., Johansson, H., Lee, K. P., Bou-Torrent, J., Stewart, K., Steel, G., et al. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10:e1004416. doi: 10.1371/journal.pgen.1004416
Wang, H., Wu, G., Zhao, B., Wang, B., Lang, Z., Zhang, C., et al. (2016). Regulatory modules controlling early shade avoidance response in maize seedlings. BMC Genomics 17:269. doi: 10.1186/s12864-016-2593-2596
Waters, M. T., and Smith, S. M. (2013). KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant 6, 63–75. doi: 10.1093/mp/sss127
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523. doi: 10.1073/pnas.1108436108
Yang, D. L., Yao, J., Mei, C. S., Tong, X. H., Zeng, L. J., Li, Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200. doi: 10.1073/pnas.1201616109
Keywords: shade avoidance syndrome, light signaling, PIFs, hormone regulation, crosstalk
Citation: Yang C and Li L (2017) Hormonal Regulation in Shade Avoidance. Front. Plant Sci. 8:1527. doi: 10.3389/fpls.2017.01527
Received: 26 June 2017; Accepted: 21 August 2017;
Published: 04 September 2017.
Edited by:
Yunde Zhao, University of California, San Diego, United StatesReviewed by:
Yi Tao, Xiamen University, ChinaRongcheng Lin, Institute of Botany (CAS), China
Christian Fankhauser, University of Lausanne, Switzerland
Copyright © 2017 Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Li, linli@fudan.edu.cn
 Chuanwei Yang
Chuanwei Yang Lin Li
Lin Li