- Hunan Province Key Laboratory of Crop Sterile Germplasm Resource Innovation and Application, College of Life Science, Hunan Normal University, Changsha, China
Although abscisic acid (ABA) is an important hormone that regulates seed dormancy, stomatal closure, plant development, as well as responses to environmental stimuli, the physiological mechanisms of ABA response to multiple stress in rice remain poorly understood. In the ABA biosynthetic pathway, 9-cis-epoxycarotenoid dioxygenase (NCED) is the key rate-limiting enzyme. Here, we report important functions of OsNCED3 in multi-abiotic stress tolerance in rice. The OsNCED3 is constitutively expressed in various tissues under normal condition, Its expression is highly induced by NaCl, PEG, and H2O2 stress, suggesting the roles for OsNCED3 in response to the multi-abiotic stress tolerance in rice. Compared with wild-type plants, nced3 mutants had earlier seed germination, longer post-germination seedling growth, increased sensitivity to water stress and H2O2 stress and increased stomata aperture under water stress and delayed leaf senescence. Further analysis found that nced3 mutants contained lower ABA content compared with wild-type plants, overexpression of OsNCED3 in transgenic plants could enhance water stress tolerance, promote leaf senescence and increase ABA content. We conclude that OsNCED3 mediates seed dormancy, plant growth, abiotic stress tolerance, and leaf senescence by regulating ABA biosynthesis in rice; and may provide a new strategy for improving the quality of crop.
Introduction
Abscisic acid (ABA) is a sesquiterpenoid that plays a critical role in seed dormancy, stomatal closure, plant development, and biotic/abiotic stress tolerance during the plant life cycle (Nambara and Marion-Poll, 2005). Furthermore, the ABA levels in plant tissues are usually dynamic in order to respond to physiological changes and environmental stimuli. For instance, in relation to shoot and root growth, ABA is generally considered to be an inhibitor in well-watered condition, but normal ABA levels are necessary to maintain root growth (in vp5 or vp14 mutants) under water stress, and normal ABA levels are necessary for shoot development, particularly leaf expansion (in flc or aba2 mutants) under well-watered conditions (Sharp et al., 2000; Sharp and LeNoble, 2002; LeNoble et al., 2004). Under water stress, ABA is dramatically increased, and it regulates stomata closure in plants to reduce water loss. This is an ABA-dependent mechanism, which involves activating H2O2 production, which subsequently increases calcium levels in guard cells, which triggers stomatal pores closure (Tardieu and Davies, 1992; Wang and Song, 2008; Yao et al., 2013). Although the physiological importance of ABA in plant growth and abiotic stress tolerance has been well-recognized, the molecular mechanisms of ABA response to multiple stress in rice remain poorly understood.
The endogenous concentration of ABA in plant tissues is regulated by ABA biosynthesis (Ng et al., 2014). ABA is produced de novo in the ABA biosynthesis pathway, which originates from the catalysis of carotenoid precursors for several enzymes found in higher plants (Xu et al., 2013). To date, most ABA biosynthesis genes have been discovered and cloned, including those for zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NCED), and abscisic aldehyde oxidase (AAO) (Taylor et al., 2000; Hauser et al., 2011). The first committed step of ABA biosynthesis is the cleavage of 9-cis-violaxanthin or 9-cis-neoxanthin by NCED to produce xanthoxin (C15) (Tan et al., 1997; Milborrow, 2001). The first NCED gene to be identified and cloned was VP14 in maize (Schwartz et al., 1997), and subsequently, NCED genes were isolated from other plant species (Priya and Siva, 2015) such as tomato (Burbidge et al., 1999), avocado (Chernys and Zeevaart, 2000), Arabidopsis (Rock and Zeevaart, 1991; Tan et al., 2003), Malus (Xia et al., 2014), and Brassica napus (Xu and Cai, 2017).
Previous studies have shown that increased NCED transcript levels could promote ABA biosynthesis and increase ABA accumulation in plants (Qin and Zeevaart, 2002; Martinez-Andujar et al., 2011). Several NCED mutants have been identified and studied; many have reduced resistance to severe environmental conditions or abnormal and defective morphology. The VP14 from maize is expressed in embryos and roots and is strongly induced in leaves by water stress. VP14 from maize is responsible for promoting seed dormancy and water stress resistance by controlling ABA levels in plants (Tan et al., 1997; Sharp and LeNoble, 2002). In addition, BnNCED3 was isolated from Brassica napus, and ectopically expressed in Arabidopsis. The authors found that overexpression of BnNCED3 elevated ABA levels in plants, delayed seed germination, reduced lateral root initiations, and promoted leaf senescence and an early flowering time (Xu and Cai, 2017). Furthermore, NCED is a multigene family; there are five NCED members confirmed in Arabidopsis, each of which is located in specific tissues, where they control ABA biosynthesis and regulate development (Tan et al., 2003). However, many of these proteins share redundant functions (Finkelstein, 2013). AtNCED6 is constitutively expressed in the endosperm (Lefebvre et al., 2006; Martinez-Andujar et al., 2011), but AtNCED9 is expressed both in the embryo and endosperm during seed development (Toh et al., 2007; Seo et al., 2016). AtNCED5 is also expressed in the seed at later periods of development. AtNCED5, AtNCED6, and AtNCED9 co-regulate seed development and dormancy (Frey et al., 2012). AtNCED3 is predominantly induced by water stress and controls endogenous ABA content under water stress conditions (Endo et al., 2008; Hao et al., 2009), and thus, the AtNCED3 T-DNA insertion mutant has a water deficiency-sensitive phenotype (Iuchi et al., 2001). AtNCED5 and AtNCED3 participate together in the water stress response in plants; moreover, nced3 and nced5 mutants suppress vegetative growth of Arabidopsis (Frey et al., 2012).
To date, five NCED genes have been found and implicated in ABA biosynthesis in rice (Zhu et al., 2009). Gene expression analysis showed that OsNCED1 has the highest expression level in rice leaves, and acts as the housekeeping gene under normal conditions, but it is significantly suppressed by water stress (Ye et al., 2011). The expression levels of OsNCED2 and OsNCED3 are both related to delayed seed germination (Zhu et al., 2009; Song et al., 2012). Additionally, OsNCED3, OsNCED4, and OsNCED5 are induced by water deficiency stress (Teng et al., 2014; Zhang et al., 2015). Although OsNCED3 was ectopically expressed in Arabidopsis, and contributes to increased ABA accumulation, enhanced drought tolerance, and altered leaf morphology (Hwang et al., 2010), the native function of OsNCED3 in rice is still unclear. In this study, we investigated the physiological roles of OsNCED3 using the knock-out nced3 mutants and OsNCED3-overexpressing transgenic rice plants. Our results suggest that OsNCED3 regulates seed dormancy, stomata aperture, plant growth, abiotic stress tolerance, and leaf senescence by altering ABA accumulation in rice.
Materials and Methods
Mutant Generation Methods
CRISPR/Cas9 system (provided by Professor Lijia Qu) was applied to generate nced3 mutants. The OsNCED3 coding sequence was selected to guide RNA design. According to NCBI blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and CRISPR GE tool (http://skl.scau.edu.cn/) analysis, the specific 20 bp spacer sequences (GCCGCCCGCGCGCGCGCTGC) immediately before a PAM sequence(5′-NGG-3′) were selected and synthesized on the base of protocol (provided by Professor Lijia Qu) with additional BsaI restriction site sequences at the pairs of sequences ends. The double-stranded spacers were generated by annealing and inserted into the BsaI-digested pOS-sgRNA vector. Then this constructed pOS-sgRNA was implemented with the destination vector pH-Ubi-Cas9 through gateway cloning LR reaction (Invitrogen, Shanghai, USA). The resulting plasmids were sequenced.
The completed pH-Ubi-Cas9 constructs were then transformed into rice (O. sativa L. japonica) by Agrobacterium-mediated. The transgenic seedlings were grown in a chamber at 28°C under a 14 h light/10 h dark cycle or in a greenhouse with natural sunlight. For mutation detection, genomic DNA was extracted from whole mutant seedlings by using PlantGen DNA kit (CWbiotech, Beijing, China). Genomic DNA was amplified by using mutation detection primers which designed to flank the designated target site. Then, the sequence chromatograms were analyzed by DSDecode tool (http://dsdecode.scgene.com/) to check the genotype of the transgenic plants. The 35 T0 hygromycin-resistant transgenic plants have three genotypes including homozygous, bi-allelic and heterozygous lines. The two independent homozygous mutant lines from the T1 generation which were named nced3-1 and nced3-2 used for future study. The primers used for CRISPR/Cas9 (U3-NCED3-F/R) and mutation detection (NCED3-J-F/R) are listed in Table S3.
The transgenic plants off-target effects analysis was performed on the base of CRISPR GE tool (http://skl.scau.edu.cn/). The potential off-target sites were detected by using site-specific genomic PCR and sequenced to determine whether the potential off-target sites were also edited. The site-specific primers are listed in Table S3.
Plant Growth Condition and Treatment
All of the experiments were performed using O. sativa L. japonica seeds from the same harvest and storage conditions. Seeds were surface-sterilized in 75% (v/v) ethanol for 2 min and rinsed twice in sterile double-distilled water, and then in 2.5% (v/v) NaClO for 30 min. The seeds were then rinsed five times in sterile double-distilled water. The sterilized seeds were soaked in water at 30°C for 2 d and then germinated for 3 d at 28°C. Seedlings were grown in Hoagland's culture solution as described previously (Li and Cheng, 2015). Plants were cultured in a growth chamber at 28°C under a 14 h light/10 h dark photoperiod. For abiotic stress, three-leaf stage seedlings were placed into solutions with final concentrations of 25% PEG6000 (−0.51 Mpa, the solution was aerated in the experimental system), 150 mM NaCl and 100 mM H2O2. All treatments were repeated more than three times. The seedlings were harvested for RNA isolation or phenotype analyses.
For drought treatments at the seedling stage, the germinated seedlings were transplanted into a soil mix (2:1 soil:vermiculite) medium in oblong white resin pots (L33 × W19 × H14 cm). The soil was river alluvial paddy soil collected from experimental field of the Hunan Normal University, Changsha, 28°11′49″N, 112°58′42″E, Hunan province, China. The seedlings were grown to the three-leaf stage and then were subjected to withholding water for 15 d and photographed, followed watering was resumed for 7 d. The plants were photographed again and the number of surviving plants was scored. During the reproductive-stage water stress, the seedlings were grown in potted bucket (40 cm diameter, 40 cm height) until the inflorescence meristem appeared; then we withheld watering for 7 d until the leaves rolled. We photographed the plants at this time and then resumed watering to the mature stage and statistically assessed the seed set rate.
Seed Germination Analysis
The harvested seeds from nced3 mutants and WT rice plants were surface-sterilized and directly sown onto the sterile filter papers for a seed germination assay. Seeds were placed in a Polymethyl methacrylate germinating box and cultured in growth chamber (ZHUJIANG, Guangzhou, China) keep at 30°C. The germination ratio was based on radicles over 1 mm and was recorded after 2 d of imbibition. Moreover, we determined the shoot and root length. Three independent experiments were performed for this analysis.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from rice seedlings or specific tissues at reproductive stage using Trizol according to the manufacturer's instructions (Invitrogen, Shanghai, USA). Approximately 1 μg of total RNA was used for first-strand cDNA synthesis with the Synthesis Kit (Thermo Fisher Scientific, USA). The reaction products were diluted ten-fold and used as the template for real-time PCR with three biological replicates using the SYBR Premix Ex Taq kit (Takara, Dalian, China). qRT-PCR was performed with an Applied Biosystems Quant Studio 5 (Thermo Fisher Scientific, USA) real-time PCR System. The gene-specific primers used in the qRT-PCR analysis are listed in Table S3.
Generation of OsNCED3 Overexpression Lines
The coding sequence of OsNCED3 was amplified by the pHB-NCED3-F/R primers (Table S3) and then inserted into pHB binary vector behind 2 × 35S promoter. The recombinant construct pHB-35S::OsNCED3 was transformed into calli of O. sativa L. japonica through Agrobacterium mediated transformation. The T2 generation transgenic seeds were screened through planted on MS medium containing 50 μg/L hygromycin (Hgr) for 7 d. The 100% resistance to Hgr were selected as homozygous lines.
Gus Staining and Subcellular Localization Assay
For histochemical analysis, the OsNCED3 promoter, a 2.27 kb genomic DNA fragment upstream region from the transcriptional initiation site was amplified by PCR using the primers 1301-OsNCED3-F/R (Table S3) and fused into pCambia1301 vector to drive the reporter gene β-glucuronidase (GUS). The ProNCED3-GUS construct was transformed into rice (O. sativa L. japonica). The ProNCED3-GUS transgenic plants (T1) were used to histochemical staining analysis. Histochemical staining was conducted as previously described (Li et al., 2015). The tissues of ProNCED3-GUS transgenic plants (T1) were incubated in the GUS staining solution (50 mM sodium phosphate at pH 7.0, 10 mM EDTA, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 10% methanol, 0.1% Triton X-100, and 1 mg mL−1 of X-Gluc (Sangon, Shanghai, China) dissolved in N,N-dimethyl formamide) at 37°C for 10 h, the samples were incubated in 75% (v/v) ethanol to clear the chlorophyll. The chlorophyll-free samples were observed and imaged with OLYMPUS SZX7 and BX51 microscope.
To investigate the subcellular location of OsNCED3, the coding sequence for OsNCED3 was amplified and ligated into the pEZS-eGFP vector. The recombinant vector pEZS-OsNCED3-eGFP was transformed into Arabidopsis protoplasts according to a protocol described previously (Meng et al., 2014). The GFP fluorescence in the transformed protoplasts was visualized with a confocal fluorescence microscope (Nikon A1, Japan).
Analyses of Proline Content, Electrolyte Leakage, and Water Loss Rate
The proline content determined as previously described (Bates et al., 1973). Approximately 0.5 g leaf segment samples from transgenic and WT plants were homogenized in 6 mL of 3% aqueous sulfosalicylic acid and centrifuged at 4,000 × g for 10 min. Then, 2 mL of supernatant was incubated with 2 mL of acid ninhydrin and 2 mL of glacial acetic acid in boiling water for 40 min, and cooled on ice to stop the reaction. For every reaction, 4 mL toluene was added and the reaction solution was oscillated for 30 min at room temperature. The absorbance was measured at 520 nm with a spectrophotometer.
The relative electrolyte leakage measurements were collected as previously described (Lou et al., 2017) with slight modification. The leaves were placed in tubes containing 20 mL of deionized water and then placed under a vacuum for 1 h. Samples were then sealed and placed at 28°C in an incubator for 5 h. The water conductance was measured with a conductivity meter.
Water loss rates were measured in detached leaves placed in dishes in the light at room temperature. Fresh weight was measured at set times from 0 to 7 h. The proportion of the initial fresh weight represented the water loss rate.
DAB Staining and Superoxide Dismutase (SOD) and Hydrogen Peroxidase (CAT) Activity Assay
H2O2 staining in leaves was performed following a previously described protocol (Hu et al., 2005). Detached leaves were infiltrated with 1 mg mL−1 3,3′-diaminobenzidine (DAB) solution (contain 0.05% Tween and 10 mM Na2HPO4, pH 3.8) at 25°C for 8 h in the dark. Leaves were then immersed in 96% boiling ethanol for 15 min to disrupt the chlorophyll. The samples were transferred to fresh ethanol and photographed. The SOD and CAT were extracted from 0.2 g leaf samples by grinding them in 2 mL of 50 mM phosphate buffer (pH 7.0) at 4°C. SOD and CAT were determined using commercial assay kits purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China), following the manufacturer's instructions. The absorbance of SOD and CAT was measured at 550 and 240 nm, respectively. All measurements were similar in all three individual replicates.
Analysis of Rice Stomata by Scanning Electron Microscopy
Leaves from wild type and nced3 mutant plants (1 month old) were treated with water stress or 100 mM H2O2 and then immediately fixed by 3% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at 4°C for 5 h, followed by washing twice with 0.1 M phosphate buffer for 10 min. Subsequently, these samples were fixed by 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) at 4°C for 2 h, washed twice with 0.1 M phosphate buffer for 10 min, dehydrated serially in 40, 50, 60, 70, 80, 90, and 100% ethanol solution for 20 min each, and then dried by critical-point dryer (Zhang et al., 2011). The samples were coated with gold and monitored by scanning electron microscopy (SU8010, Hitachi, Japan).
Measurement of Chlorophyll
Chlorophyll was extracted from 0.15 g of rice flag leaf tissue with 80% ice-cold acetone, and its content was measured according to the absorbance at 663 nm and 645 nm with a UV2400 UV/VIS spectrophotometer based on the protocol described by Lichtenthaler (1987).
Measurement of ABA Content
Leaf tissues (300 mg) were ground in liquid nitrogen. The powder was extracted with 1.5 mL of extraction solution (methanol:H2O:methanoic acid in a ratio of 7.9:2:0.1) and kept overnight at 4°C. The samples were centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was collected, dried under nitrogen gas and dissolved in 2 mL of 0.1 M ammonia solution. Crude extracts were purified using a MAX column that was pre-treated with 2 mL methanol and then with 2 mL 0.1 M ammonia solution. After the supernatant was loaded onto the MAX column, the column was washed with 2 mL of 0.1 M ammonia solution and followed by 2 mL of methanol. The ABA was eluted with 4 mL of methanol containing 1% formic acid. The eluent was dried under nitrogen gas and dissolved in 0.2 mL of methanol, and then filtered through a 0.2 μm nylon membrane. [2H6]ABA was used as internal standard. ABA levels were quantified using liquid chromatography-tandem mass spectrometry (LC–MS/MS) with three biological replicates.
Statistical Analysis
All experiments were repeated in at least three biological replicates for each treatment. Data were considered statistically significant at a P < 0.05 using Student's t-test. Data are the mean ± SD from three independent experiments.
Results
OsNCED3 Expression Pattern and Protein Localization
To observe the temporal and spatial expression patterns of OsNCED3 in various rice plant tissues, we used the 2.27 kb upstream region from the OsNCED3 transcriptional initiation site as the promoter to drive the reporter gene β-glucuronidase (GUS). Immunohistochemical staining of transgenic plants expressing ProNCED3-GUS showed that OsNCED3 was expressed in all of the tissues we tested, which included embryo, coleoptile, root, leaf, culm, node, flower, stigma, and pollen, with especially high expression in flowers and roots (Figure 1A). Quantitative RT-PCR (qRT-PCR) analysis also showed that OsNCED3 transcripts were detected in above tissues and the highest OsNCED3 expression in flowers and roots (Figure 1B). In addition, OsNCED3 expression levels were significantly upregulated under NaCl, and PEG treatment for 1 h. We also found that H2O2 could rapidly induce the increase of OsNCED3 transcript levels (Figure 1C). Additionally, GUS activity significantly increased in leaves, sheaths and roots under dehydration, NaCl and H2O2 stress, respectively (Figure S1). These results suggest that OsNCED3 could play a role in tolerance to multiple types of abiotic stress in rice.
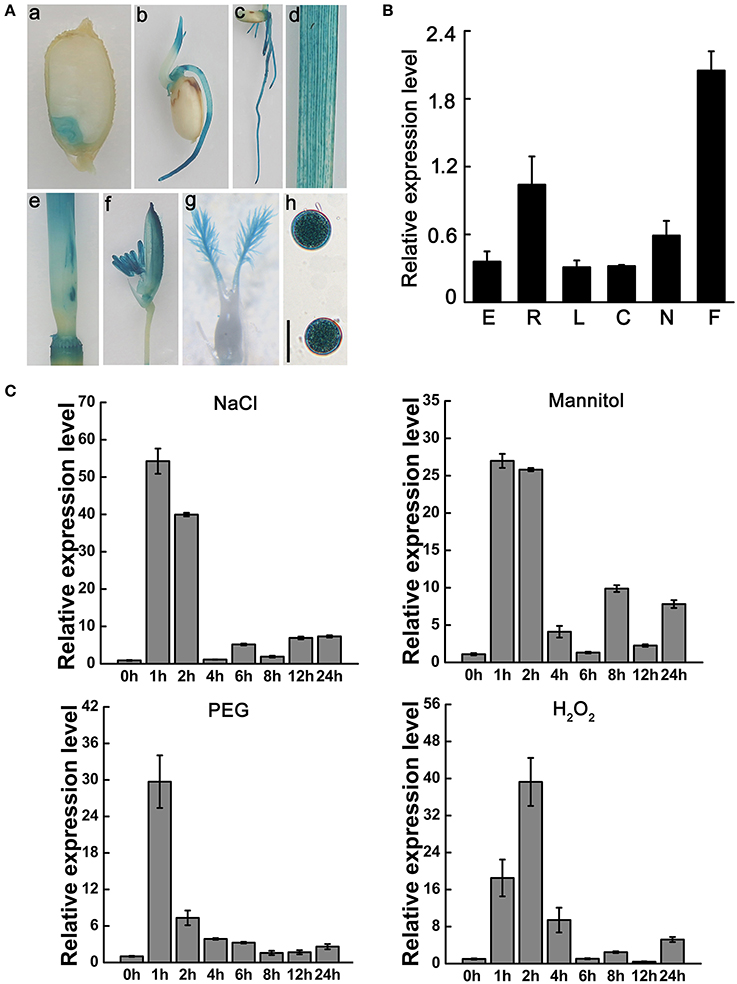
Figure 1. Expression analysis and subcellular localization of OsNCED3. (A) Histochemical staining of ProNCED3-GUS transgenic plants, including mature seed (a), bud sheath (b), root (c), leaf (d), culm and node (e), flower (f), stigma (g), pollens(h). Bars = 20 μM. (B) Transcription level of OsNCED3 in different organs. E(embryo), R(root), L(leaf), C(culm), N(node), F(flower). (C) OsNCED3 expression in abiotic stress by qRT-PCR. The three-leaf stage seedlings were performed to 150 mM NaCl, 250 mM Mannitol, 25% PEG and 100 mM H2O2 treatment, respectively. The seedling samples were collected for OsNCED3 expression analysis. Data are the mean ± SD for three replicates.
Previous studies reported that 9-cisepoxycarotenoid dioxygenase (NCED) oxidative cleave cis-violaxanthin and cis-neoxanthin into xanthoxin was occurred in Chloroplast and other plastid (Ye et al., 2012). To identify the subcellular localization of OsNCED3, we constructed an 35S::NCED3-eGFP fusion protein and transiently transformed Arabidopsis protoplasts. Confocal microscopic visualization demonstrated that the green fluorescent signal of NCED3-eGFP co-localized with the autofluorescence of the chlorophyll, which showed that OsNCED3 was targeted to chloroplasts (Figure 2).
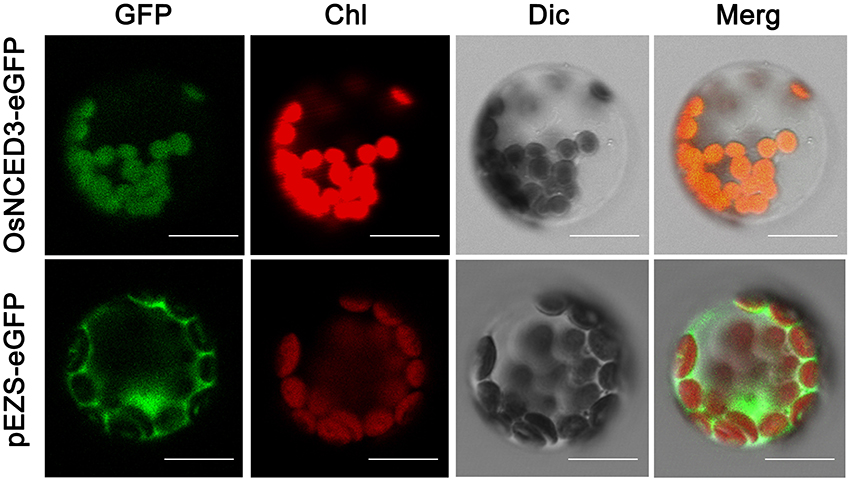
Figure 2. Subcellular location of OsNCED3 proteins. OsNCED3-eGFP fusion protein transiently expressed in Arabidopsis chloroplast and visualized by confocal microscopy, Bars = 20 μm.
OsNCED3 Regulates Seed Dormancy and Post-Germination Plant Growth in Rice
OsNCED3 is constitutively expressed in various tissues, and is significantly induced by multiple abiotic stressors. To investigate the native OsNCED3 function in rice plant growth and abiotic stress, we used the CRISPR/Cas9 system to express pCRISPR-NCED3 constructs, and transformed rice with Agrobacterium tumefaciens-mediated transformation. We obtained 35 hygromycin-resistant transgenic plants, including 32 nced3 mutants. Sequence analysis indicated that 22.9, 3.7, and 64.8% of these 35 transgenic plants were homozygous, bi-allelic and heterozygous lines, respectively (Table S1). Two independent homozygous lines (nced3-1 and nced3-2) were used for further study. The nced3-1 mutant has a 54 bp deletion that results in an 18-amino-acid deletion in the protein, and the nced3-2 mutant has a one-nucleotide insertion results in a frameshift mutation that promotes early termination of protein translation (Figure 3A, Figures S2, S8).
To confirm that the OsNCED3 mutant phenotypes were not an off-target effect of CRISPR/Cas9 editing, we found seven loci with the highest ranking for off-target potential (Table S2) based on the prediction of the CRISPR-GE tool, and designed primers to amplify these regions. As shown in Figure S3, no mutations were detected at the potential off-target loci in the rice genome in these obtained mutants lines.
To investigate whether OsNCED3 expression affects rice seed germination, the freshly harvested seeds were used for comparison of wild type and nced3 germination rates through counting at 7 days after soaking. The germination rates of nced3-1 and nced3-2 mutants were slightly higher than those of the wild type (Figure 3B). In addition, the post-germinated nced3 mutants exhibited earlier growth of roots and shoots compared to wild type seedlings, and the roots and shoots of both nced3 mutants were significantly longer than those of the wild type (Figures 3C–E). At the three-leaf stage, nced3-1 and nced3-2 mutant roots and shoots were still longer than those of the wild type (Figures 3F,G). However, the mutants had no obvious differences after the tillering stage compared to the wild type when the plants were grown after this same developmental stage (data not shown). These results indicate that knock-out of OsNCED3 may altered plant growth in seedling stage.
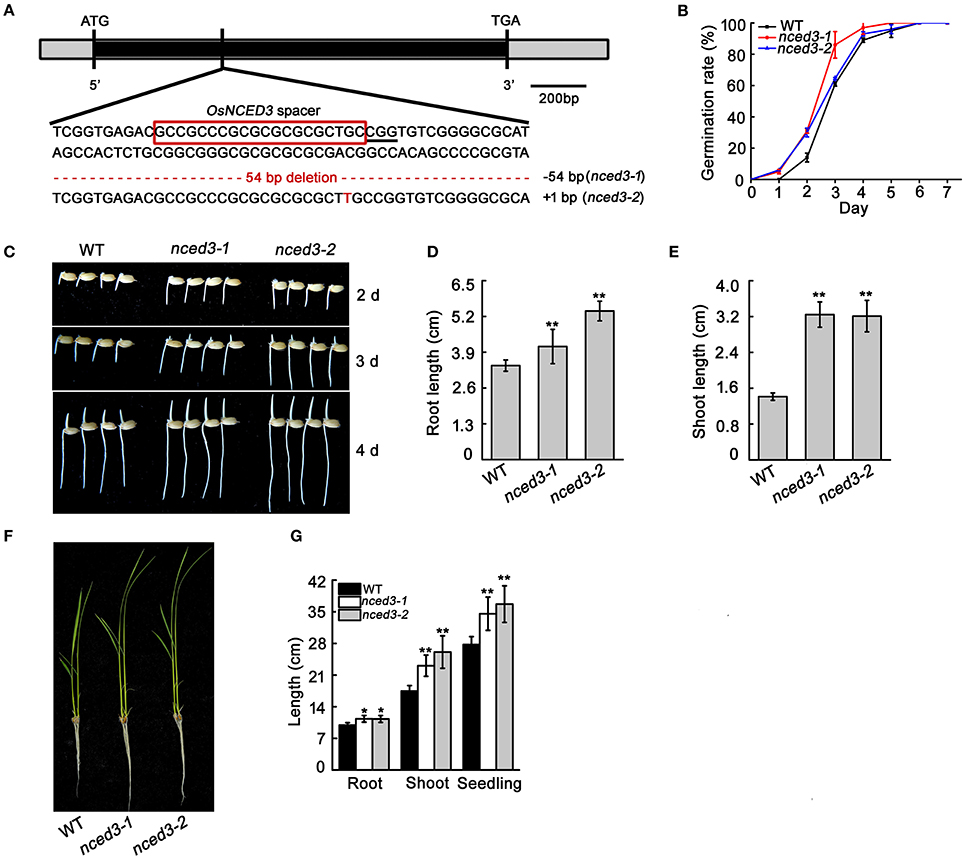
Figure 3. CRISPR/Cas9 induced mutation of OsNCED3 and the phenotype of seed germination and early seedling growth in nced3 mutants. (A) Targeted mutagenesis of the OsNCED3 gene. The OsNCED3 mutation site is shown on the gene structure. The sequence in the red box represents the OsNCED3 spacer, and PAM is labeled by underline. The indels are shown in red dashes or letters. (B) Germination rate of wild type and nced3 mutants. Data are the mean ± SD for three replicates (each replicate containing 100 seeds). (C) Growth state at post-germination 2 d, 3 d, and 4 d for wild type and nced3. (D) Root and (E) shoot length of plants post-germination 4 d. (F) Seedling characteristics in the three-leaf stage. (G) Length measurement of (E) seedlings. Data shown are ± SD (n = 14). Similar results in three individual experiments. Asterisks indicate statistically significant differences (Student's t-test; *P < 0.05, **P < 0.01).
Compared to Wild-Type Rice, nced3 Mutants Are More Sensitive to NaCl, PEG, and Oxidation Stress
Our qRT-PCR analysis showed that OsNCED3 expression was noticeably induced by NaCl and PEG stress. Therefore, we treated 2-week-old nced3 mutant and wild type seedlings with Hoagland's nutrient solution that contained 150 mM NaCl or 25% PEG. After 5 days of NaCl stress, more leaves of the two nced3 mutants had withered compared to the wild type plants. After the high-salt treatment, the plants were transferred to normal nutrient conditions to recover for 5 d. The recovery was 48% for nced3-1 and 32.7% for nced3-2, which was lower than the 66.6% observed for the wild type (Figures 4A,B). Furthermore, 2-week-old nced3 mutants and wild type seedlings were subjected to 25% PEG treatments for 15 d, after which most of the nced3 mutants leaves withered, whereas the wild type showed slightly response. The seedlings were then transferred to normal conditions and allowed to recover for 7 d, but only 69.7% and 55.2% of the nced3-1 and nced3-2 seedlings survived, respectively, which were significantly lower than the 88.4% survival observed for the wild type (Figures 4A,C). These results indicate that the nced3 mutants had a decreased capacity for high salinity and osmotic stress tolerance.
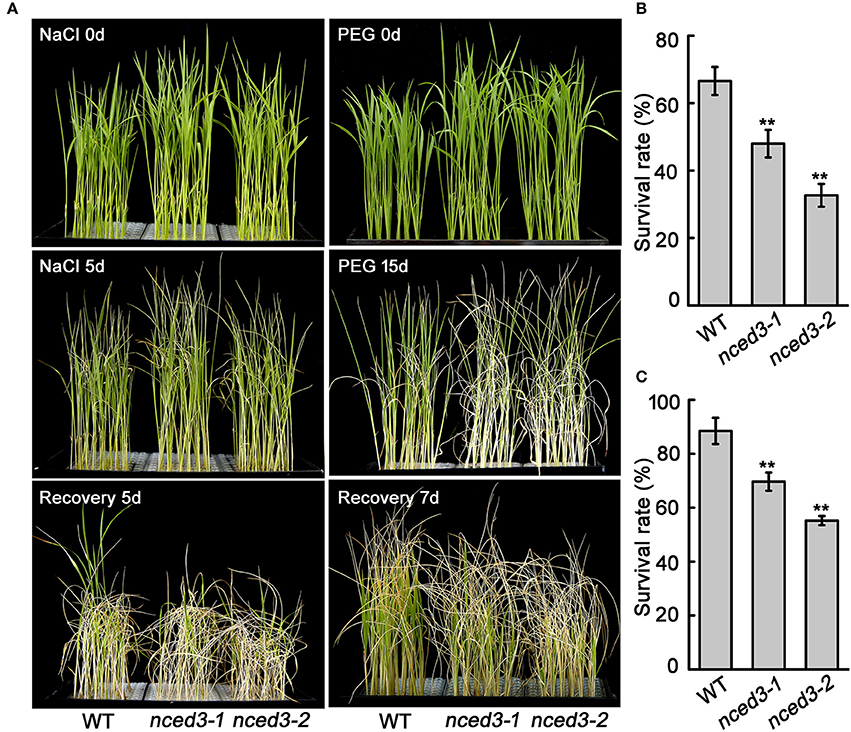
Figure 4. The nced3 mutants showed decreased salt and PEG stress tolerance. (A) The phenotype of three-leaf stage seedlings of nced3 mutants and wild type under NaCl and PEG stress. (B,C) Statistics for survival rate after salt and PEG stress. The number of surviving plants as a proportion of the total plants is shown. Data shown are ± SD from three independent replicates. Asterisks indicate statistically significant differences (Student's t-test; **P < 0.01).
OsNCED3 also responded to H2O2 treatment. To examine whether OsNCED3 confers ROS tolerance, nced3 mutants and wild type seedlings were subjected to H2O2 treatment for 2 d at the three-leaf stage. Whereas almost all of the nced3 leaves had wilted under these conditions, wild type leaves were generally unaffected (Figure 5A). Using DAB staining, we detected no differences in the untreated leaves of nced3-1, nced3-2 and wild type rice, whereas the leaves of nced3-1 and nced3-2 plants had dark brown discoloration compared to wild type leaves, which only had a few brown regions at the leaf apexes (Figure 5B). These observations suggest that OsNCED3 loss-of-function mutant plants were susceptible to H2O2 stress. H2O2 is a vital stress signaling molecule in plants, and can increase the superoxide dismutase (SOD) and hydrogen peroxidase (CAT) activity in leaves (Hu et al., 2005). To determine whether nced3 mutants had altered antioxidant enzyme activity during H2O2 treatment, SOD and CAT activity was measured. The activity of these two antioxidant enzymes in nced3 mutants and wild type plants were enhanced during H2O2 treatment as compared to control conditions, but the SOD and CAT activity of nced3 mutants was lower than that of wild type seedlings under H2O2 stress (Figure 5C). These results indicate that OsNCED3 could promote antioxidant enzyme activity when plants are exposed to oxidative damage.
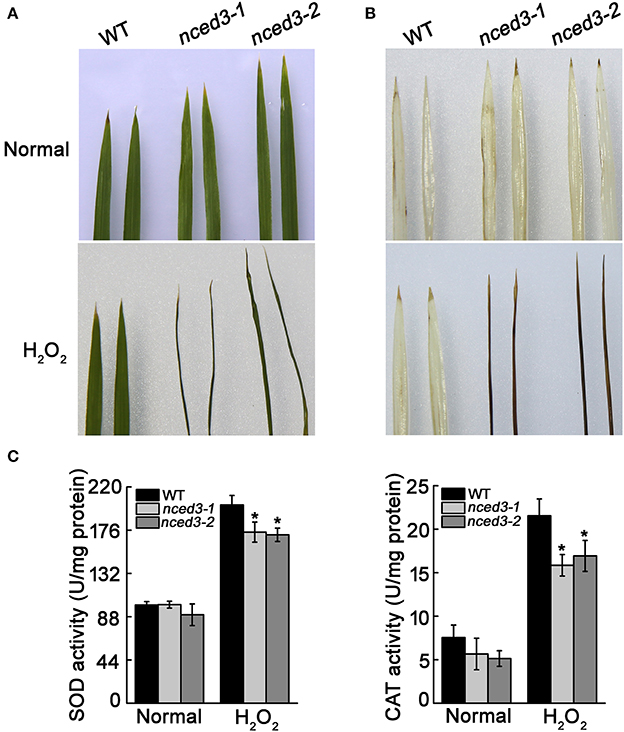
Figure 5. Analysis of H2O2 accumulation and oxidative enzymes under H2O2 stress condition. (A) Phenotype of nced3 and wild type leaves after 100 mM H2O2 treatment for 2 d. (B) DAB staining of H2O2 accumulation. Three independent replicates were performed. (C) SOD and CAT activities of nced3 and wild type three-leaf-stage seedlings under 100 mM H2O2 treatment for 24 h. Data shown are ± SD from three independent replicates. Asterisks indicate statistically significant differences (Student's t-test; *P < 0.05).
nced3 Mutant Rice Had Decreased Water Stress Tolerance and Stomata Closure
The nced3 mutants and wild type seedlings were grown to the three-leaf stage in soil before watering was withheld for 15 d, and then watering was resumed for 7 d. Approximately 83.2% of the wild type plants recovered from this water stress treatment, but only 40.2 and 20% of the nced3-1 and nced3-2 plants recovered, respectively (Figures 6A,B). The water stress sensitivity of the nced3 mutants was also tested at the reproductive stage. We observed that 76.7 and 85.9% of the nced3-1 and nced3-2 mutant leaves rolled, respectively, which was more than was observed in the wild type (35.3%) under water stress (Figures S5A–C). After water stress, the seed set rates of nced3-1 and nced3-2 mutants were only 55.8 and 47.7%, which was distinctly lower than the 70.7% seed set rate measured in the wild type (Figures S5D,E).
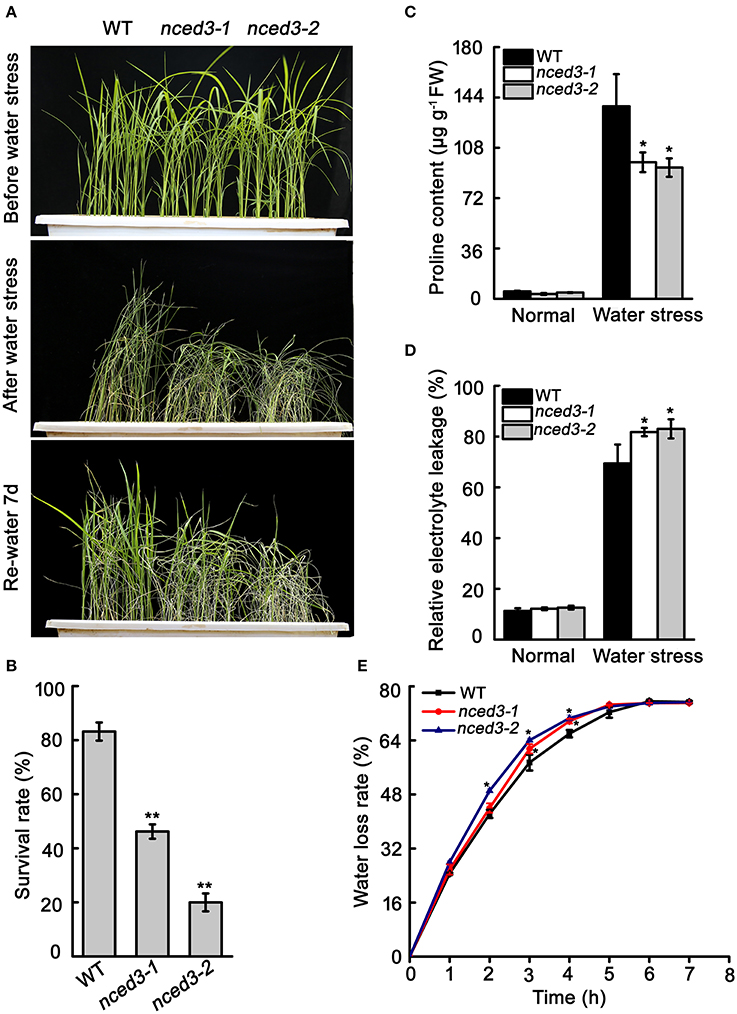
Figure 6. Water stress tolerance of nced3 seedlings. (A) Phenotype of nced3 in water stress. (B) Survival rate after drought stress. The number of surviving plants as a proportion of the total plants is shown. (C, D) Proline and relative electrolyte leakage determination after water treatment. Data shown are ± SD from three independent replicates. (E) Measurement of water loss rate in wild type, nced3-1, and nced3-2 seedlings. Values are the mean ± SD (n = 6 plants). Similar results were obtained from three independent replicates. Asterisks indicate statistically significant differences (Student's t-test; *P < 0.05, **P < 0.01).
Given that the altered physiological mechanism possibly responsible for water stress adaption. we examined proline content and relative electrolyte leakage in leaf tissues. Under normal conditions, these two factors were very low, and there were no differences between the wild type and nced3 mutants. After 10 d of water stress, proline content in leaves significantly increased in the wild type and the two nced3 mutants; however, the nced3 mutants leaves accumulated less proline than the wild type (Figure 6C). At the same time, relative electrolyte leakage was greater in nced3 mutants than in the wild type under drought stress (Figure 6D). We also evaluated water loss avoidance in nced3 and wild type plants by water loss rate analysis. The detached leaves of nced3 mutants lost water faster than those of the wild type (Figure 6E). These results indicate that OsNCED3 plays a vital role in water stress tolerance in rice.
To further analyze that the water loss rate in nced3 mutants were more faster than wild type. We investigated the stomatal aperture and density in nced3-2 mutant and wild type plants. Under normal conditions, the percentages of completely open stomata in the nced3-2 mutant plants were little higher than wild type plants, but other two-type stomata have no difference between nced3-2 mutant and wild type (Figures 7A,B). However, under water stress conditions, 66.1% of stomata were completely closed in wild type plants, while only 43.6% were completely closed in nced3-2 mutant plants. Additionally, only 26.1% of stomata were partially open in wild type plants, but 45.7% were partially open in nced3-2 mutant plants. Furthermore, 10.7% of stomata were completely open in nced3-2 mutant plants, and no significant difference was observed compared to the 7.8% of completely open stomata in wild type plants (Figure 7A,B). We also observed stomatal density, and confirmed that there were no obviously differences between nced3-2 mutant and wild type plants (Figure 7C). These results indicate that OsNCED3 controls stomata movement under water stress conditions.
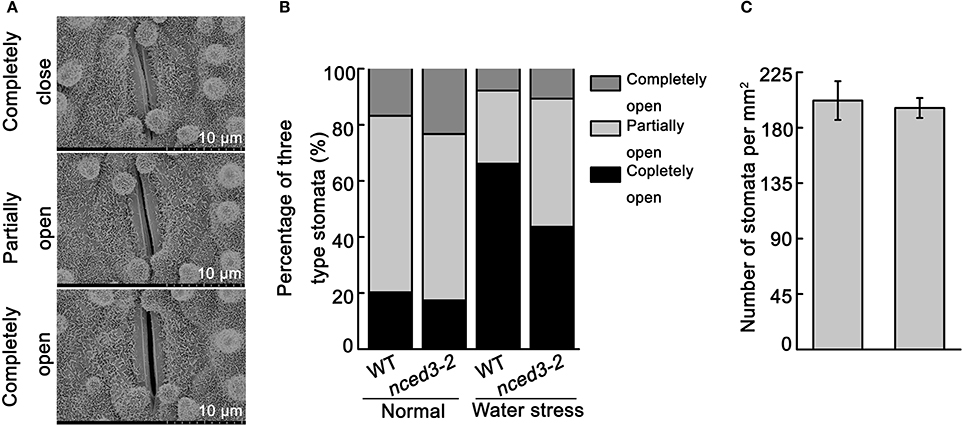
Figure 7. Stomata closure of nced3 mutant under water stress. (A) Scanning electron microscopy images of three levels of stomatal apertures, Bars = 10 μm. (B) The percentage of three levels of stomatal apertures in nced3-2 mutant and wild type plants were calculated under normal and water stress condition (n = 105 stomata for wild type; n = 98 stomata for nced3-2 mutant). (C) Stomata numbers per mm2 were counted in middle leaves from nced3-2 mutant and wild type plants, respectively. Data shown are ± SD from there independent replicate.
OsNCED3 Affects ABA Accumulation and ABA-Related Gene Expression
ABA was well described as regulates plant growth and enhances water stress adaption, and NCED catalyzes the key regulatory step in ABA biosynthesis (Ng et al., 2014). Hence, we measured the ABA content of shoots and roots at the three-leaf stage, and found that the ABA levels in the shoots of mutants (5.5 ng/g for nced3-1 and 6.2 ng/g for nced3-2) and roots of mutants (1.7 ng/g for nced3-1 and 1.4 ng/g for nced3-2) were both noticeably lower than that in wild type plants (10.2 ng/g in shoots and 3.2 ng/g in roots) (Figure 8A). Therefore, nced3 mutants had longer shoots and roots than wild type plants, possibly due to a decrease in ABA content. Moreover, the ABA levels in the leaves of nced3 mutants and wild type were also investigated under normal and water stressed growth conditions. Under normal conditions, ABA levels in the nced3 mutants (6.3 ng/g for nced3-1 and 6.6 ng/g for nced3-2)were approximately 50% of that in wild type (13.3 ng/g). After the water stress treatment, ABA content in nced3 and wild type seedlings significantly increased, but the wild type (1429.4 ng/g) ABA content was 2.3 times higher than nced3 mutants (650.9 ng/g for nced3-1 and 601.9 ng/g for nced3-2) (Figure 8B). Therefore, the increased water stress sensitivity in the nced3 mutant seedlings correlates with the mutation in OsNCED3, which could reduce endogenous ABA biosynthesis.
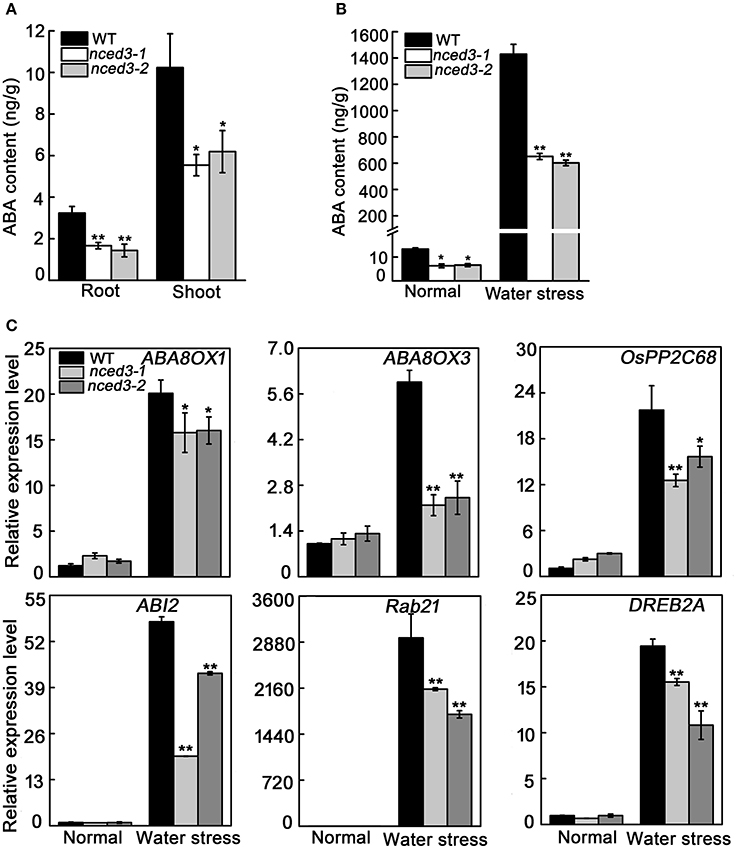
Figure 8. ABA accumulation and ABA-related gene expression in nced3 mutants. (A) The shoot and roots ABA content in normal condition, (B) The leaf ABA content under water stress, (C) The leaf ABA-related genes expression in wild type and nced3 mutants three-leaf stage seedlings under water stress for 5 d. Data shown are ± SD from there independent replicate. Asterisks indicate statistically significant differences (Student's t-test; *P < 0.05, **P < 0.01).
We further determined the transcript levels of ABA catabolic genes and ABA signaling pathway genes under water stress. Consistent with the increase in ABA levels after water stress treatment, expression of OsABA8ox1, OsABA8ox3, OsPP2C68, OsABI2, OsRab21, and OsDREB2A genes were noticeably induced in all plants, but nced3 mutant seedlings had less of an increase compared to the expression levels measured in wild type seedlings (Figure 8C). These results suggest that OsNCED3 plays an important role in ABA biosynthesis, and indirectly up-regulates ABA-related gene transcription under water stressed growth conditions.
Overexpression of OsNCED3 Enhanced NaCl and Water Stress Tolerance and Increased Endogenous ABA Content
To confirm the function of OsNCED3 in response to NaCl and water stress, transgenic lines (OE1, OE2) of OsNCED3-overexpressing plants were obtained (Figure S6). The growth of OsNCED3-overexpressing plants were similar to wild type plants. However, OsNCED3-overexpressing plants were more tolerant to high salinity stress compared to wild type plants (Figure 9A). The survival rate of OE1 and OE2 was 64.9 and 81.6%, respectively, but the survival rate of wild type was only 54.1% (Figure 9B). Additionally, the OsNCED3-overexpressing seedlings and wild type seedlings were grown in soil and withholding water for water stress analysis. After withholding water for 18 d and re-watering, the OsNCED3-overexpressing plants had enhanced tolerance to water stress compared to with wild type (Figure 9C). Approximately 57.8% of the wild type plants recovered from water stress conditions, but 69.1 and 84.3% of the OE1 and OE2 transgenic plants recovered, respectively (Figure 9D).
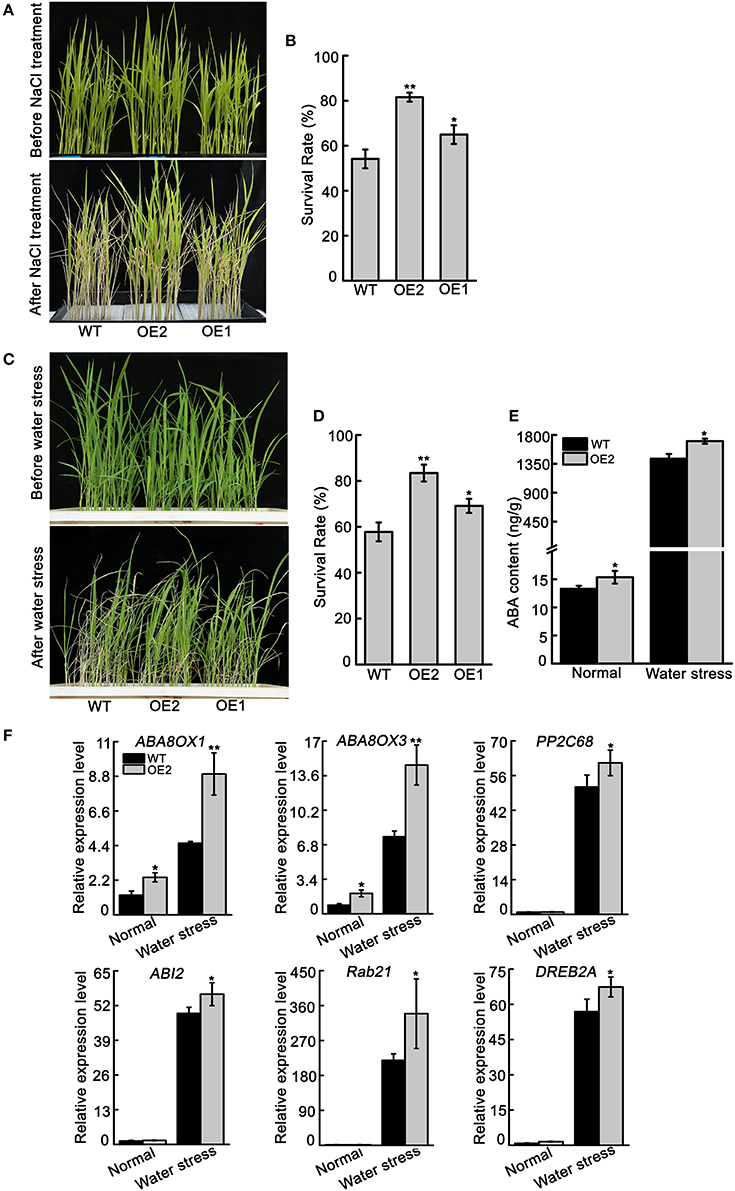
Figure 9. The phenotype of OsNCED3-overexpressing plants and ABA content and ABA-related genes transcript levels. (A) The phenotype of three-leaf stage seedlings of OsNCED3-overexpressing and wild type under 150 mM NaCl stress. (B) Statistics for survival rate after salt stress. The number of surviving plants as a proportion of the total plants is shown. (C) The phenotype of three-leaf stage seedlings of OsNCED3-overexpressing and wild type under water stress. (D) Statistics for survival rate after water stress. (E) ABA content and (F) ABA-related genes expression in OsNCED3-overexpressing and wild type three-leaf stage seedlings under water stress for 5 d. Data shown are ± SD from there independent replicate. Asterisks indicate statistically significant differences (Student's t-test; *P < 0.05, **P < 0.01).
In addition, the endogenous ABA content was measured in the leaves of OsNCED3-overexpressing and wild type plants under normal and water stress conditions. Under normal conditions, the ABA content in OsNCED3-overexpressing plants (15.4 ng/g) was higher than in wild type plants (13.3 ng/g). After water stress for 10 d, the amount of endogenous ABA accumulated in both OsNCED3-overexpressing and wild type plants, but the ABA content in OsNCED3-overexpressing plants (1702.8 ng/g) was much higher than in wild type plants (1429.4 ng/g) (Figure 9E). Consistently, the transcript levels of ABA related genes were significantly induced in OsNCED3-overexpressing and wild type plants under water stress conditions, but the expression levels were much higher in OsNCED3-overexpressing plants than in wild type plants (Figure 9F). Interestingly, the transcript levels of ABA8OX1 and ABA8OX3 were also higher in OsNCED3-overexpressioning plants than in wild type plants under normal condition, but other genes were not obviously different between OsNCED3-overexpressing and wild type plants. These results further indicate that OsNCED3 could promote endogenous ABA accumulation and enhance water stress tolerance.
OsNCED3 Promotes Leaf Senescence in Detached Rice Leaves
ABA is a positive regulator of leaf senescence (Lee et al., 2011). Dark treatment is an effective method to accelerate leaf senescence and ABA biosynthesis (Mao et al., 2017). OsNCED3 is a pivotal ABA biosynthesis gene and its transcription level in leaves is strongly induced in dark treatment compared to normal light conditions (Figure 10A). Consistent with the qRT-PCR results, β-glucuronidase activity was more abundant in leaves after 3 d of dark treatment than in leaves under control condition (Figure S7). The segments of leaves from nced3 mutants displayed minor leaf senescence in darkness, whereas OsNCED3-overexpressing in the OE1 and OE2 transgenic plants had a significant leaf senescence phenotype when compared with wild type plants under the same conditions (Figure 10B). Darkness led to significant loss of chlorophyll content in OE1 and OE2 detached leaves, but the decrease in chlorophyll content in nced3 mutants was significantly less than that in the wild type after 3 d of dark treatment (Figure 10C).
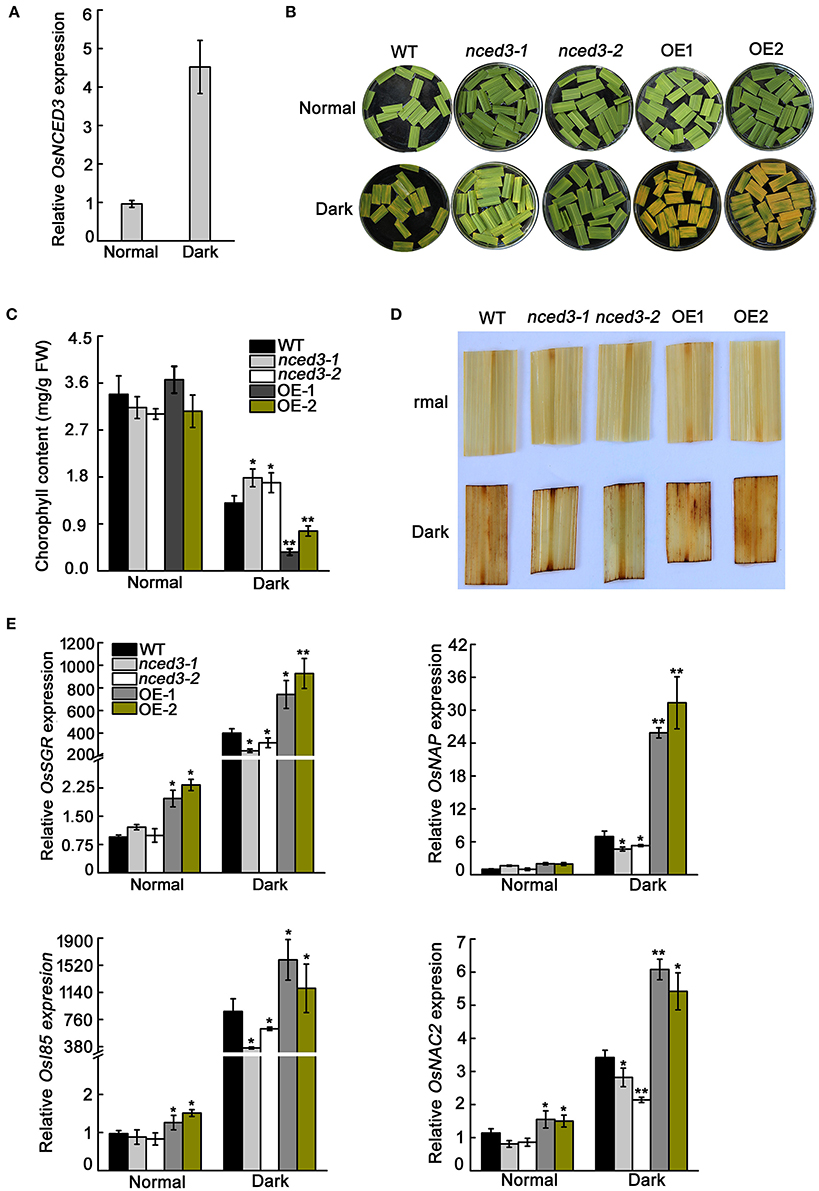
Figure 10. Dark-induced leaf senescence phenotype of leaf sections in nced3 mutant and OsNCED3-overexpressing plants. (A) Quantitative real-time PCR analysis of OsNCED3 expression after 3-day dark treatment. Data are the mean ± SD for three replicates. (B) Leaf senescence phenotype of 8-week-old nced3-1,2 and OE-1,2 plants after 3-day dark treatment. (C) Measurement of chlorophyll content in the 8-week-old nced3-1,2 and OE-1,2 leaves after dark treatment. (D) DAB staining of detached nced3-1,2 and OE-1,2 leaves after dark treatment. (E) Relative expression of OsSGR, OsNAP, OsI85, and OsNAC2 genes after 3-day dark treatment in detached nced3-1,2 and OE-1,2 leaves. Data shown are ± SD from three independent replicates. Asterisks indicate statistically significant differences (Student's t-test; *P < 0.05, **P < 0.01).
We further examined the H2O2 and senescence marker genes expression levels in darkness-treated leaves. The OE1 and OE2 detached leaves accumulated significantly higher amounts of H2O2 than wild type by DAB staining, whereas the H2O2 accumulation in nced3 mutants was lower than in the wild type (Figure 10D). As expected, transcription levels of the senescence marker genes OsSGR, OsNAP, OsI85, and OsNAC2 in OE1 and OE2 dark-treated leaves were significantly higher compared to those of the wild type, whereas these genes were showed much lower expression in the nced3 lines than in the wild type (Figure 10E). Thus, OsNCED3 is a positive regulator of ABA biosynthesis, which is related to plant leaf senescence.
Discussion
ABA is an essential hormone for seed dormancy, stomata aperture, plant development, and abiotic stress tolerance. The levels of ABA in plants is regulated by its de novo biosynthesis. NCED is the critical rate-limiting enzyme in the ABA biosynthetic pathway. OsNCED3 is a member of NCED gene family in rice, and although the partial function of OsNCED3 has been reported, its native functional characteristics in rice is still unclear. Here, we studied the functional characteristics of OsNCED3 by successfully creating nced3 mutant lines and OsNCED3-overexpressing plants.
Increased levels of ABA or inhibition of ABA degradation can maintain seed dormancy in plants (Martinez-Andujar et al., 2011). The OsNCED3 ortholog in maize, VP14, is specifically expressed in mature embryos, and its mutation resulted in nced3 mutant lines that displayed early seed germination (Figure 3B). These phenotypes were similar to the maize vp14 mutant (Schwartz et al., 2003). Other ABA biosynthesis-deficient mutants, including Osaba1 in rice (Agrawal et al., 2001) and Atnced6 and Atnced9 in Arabidopsis, contribute to seed germination (Lefebvre et al., 2006). Overexpression of ABA biosynthetic pathway genes, including NpZEP (Frey et al., 2006), TaNCED2 (Son et al., 2016), LeNCED1 (Tung et al., 2008), and GINCED1 (Zhu et al., 2007), occurs in many higher plants, and promotes seed dormancy. Thus, OsNCED3 could regulate ABA biosynthesis enzymes required for seed dormancy through increasing the ABA content in seeds.
Endogenous ABA acts as a dual modulator for plant growth and development. When excessive ABA accumulates in plant organs, it can inhibit plant development, but low endogenous ABA levels can sometimes promote plant growth (Cheng et al., 2002). The nced3 mutant lines reduced ABA content in seedlings (Figure 8A), and the shoots and roots were both significantly longer than that in the wild type (Figures 3C–G), but the growth of OsNCED3-overexpressing lines were not different from that of wild type, although ABA content was higher in OsNCED3-overexpressing lines (Figure S6 and Figure 9E). However, ectopic expression of OsNCED3 in Arabidopsis resulted in smaller and rounder leaves and midveins compared to wild type plants (Hwang et al., 2010). It is possible that OsNCED3 regulates plant development in rice or Arabidopsis through different mechanisms. Furthermore, the Arabidopsis ABA-deficient aba1 and aba2 mutants are wilted and small relative to wild type, and exogenous ABA could promote plant growth in aba1 and aba2 mutant plants (Barrero et al., 2005; Lin et al., 2007). The rice nced3 mutants displayed a different phenotype, compared to Arabidopsis ABA-deficient mutants. The higher stomata aperture in nced3 mutant possibly altered transpiration and photosynthesis, thus produced this diverse phenotype. Our results indicate that the precise modulation of ABA levels plays an important and positive role in plant growth regulation under normal environmental conditions.
The NCED genes contribute to increased ABA levels, which promotes abiotic stress tolerance in plants (Bang et al., 2013). Our study shows that expression of OsNCED3 was significantly induced by NaCl and osmotic stress (Figure 1C). Similarly, the nced3 mutants were hypersensitive to NaCl and PEG stress (Figure 4), and exhibited a significant decrease in water stress tolerance (Figure 6A). Moreover, OsNCED3-overexpressing plants had enhanced NaCl and water stress tolerance (Figures 9A–D). Several other NCED genes, including VP14 (Tan et al., 1997), AtNCED3 (Frey et al., 2012), CsNCED3 (Pedrosa et al., 2017), SgNCED (Yang and Guo, 2007), and LeNCED1 (Tung et al., 2008) were previously reported to alter water stress sensitivity if expression was inhibited or overexpressed in plants. Under water stress conditions, the plants underwent a physiological transformation to adapt to the severe environmental conditions. Proline is an osmolyte that can facilitate adaptation to water deficient conditions (Voetberg and Sharp, 1991), and electrolyte leakage is a physiological index that indicates ability of plants to resist to water stress (Jiang et al., 2016). Our results suggest that the nced3 mutants were sensitive to water stress, which can possibly be attributed to the fact that they had a low proline content and high relative electrolyte leakage (Figures 5C,D). The results were further confirmed by the enhanced water stress tolerance of OsNCED3-overexpresing plants (Figure 9C). Similar observations were observed in BnNCED3 transgenic plants (Xu and Cai, 2017). Furthermore, ABA reduce water loss by regulating stomatal closure in order to acclimate to water stress (Finkelstein, 2013). The percentage of completely close stomata in nced3 mutant is much lower than wild type plants under water stress. This is consistent with previous studies found that sapk2 mutants affect ABA-dependent stomatal movement (Lou et al., 2017). This result indicates that OsNCED3 positively regulates water stress tolerance by reducing water loss through facilitating stomata closure. Additionally, the ABA content in nced3 mutants was much lower than that measured in wild type plants under water stress conditions (Figure 8B), but OsNCED3-overexpresing plants accumulated higher ABA levels compared to wild type plants (Figure 9E) which is consistent with the observation that AtNCED3 and AtNCED5 regulate ABA accumulation under water stress (Frey et al., 2012). On the other hand, the transcription of ABA catabolic genes such as OsABA8ox1 and OsABA8ox3 (Shi et al., 2015), and signaling pathway genes such as OsPP2C68, OsABI2, OsRab21, and OsDREB2A is significantly induced to improve water stress adaptation in rice (Cui et al., 2011; Chen et al., 2014). Our study showed that the expression levels of these genes were significantly lower in nced3 mutants, but obviously higher in OsNCED3-overexprssing plants compared with wild type plants under water stress. These results indicate that OsNCED3 is responsible for ABA accumulation, and may be indirectly regulating the expression of ABA-dependent water stress-related genes under water stress.
Previous study showed that water stress-induced ABA triggered an elevation in ROS in plants to regulate stomata aperture (Yao et al., 2013); Under exogenous H2O2 treatment, the percentage of completely closed stomata in nced3 mutant was much lower than wild type plants (Figure S4), which is consistent with the observation that nced3 mutants had lower tolerance to oxidation stress than the wild type plants. Ordinarily, excessive H2O2 stimulates the dual ABA induced ROS-scavenging system to resist oxidation damage (Desikan et al., 2004; Ozfidan et al., 2012). The activity of SOD and CAT in wild type leaves was higher than the enzymatic activity measured in nced3 mutant leaves under exogenous H2O2 treatment (Figures 5C,D). This is similar to the observation that reduced expression of the OsABA8ox3 gene resulted in decreased antioxidant enzyme activity, and increased oxidative damage (Cai et al., 2015). The results for the ABA deficiency in the nced3 mutants indicate that these plants normally avoid oxidative damage by increasing ABA biosynthesis through a pathway that requires OsNCED3 gene function during oxidative stress.
ABA is a positive regulator for leaf senescence. Previous studies have shown that OsNAC2 directly binds to the promoters of the ABA metabolism-related genes OsNCED3, OsZEP1, and OsABA8ox1, which control the production of ABA in plants, and regulate ABA-dependent leaf senescence (Mao et al., 2017). Our study indicates that the expression of OsNCED3 was significantly induced by dark treatment, and leaf sections of nced3 mutants had greener leaves in the dark, but OsNCED3-overexpressing leaf sections quickly turned yellow in the dark (Figure 10A,B). Furthermore, the transcription factors OsNAC2 and OsNAP participated in the aging process dependent on ABA biosynthesis and the leaf-senescence marker genes OsI85 and OsSGR (Chen et al., 2014; Mao et al., 2017). The transcription levels of these genes in dark-treated leaves from the OsNCED3-overexpressing lines were notably higher than those in the nced3 mutants (Figure 10E). This is similar to the observation that overexpression of BnNCED3 in Arabidopsis promotes leaf senescence (Xu and Cai, 2017). Consequently, our results further confirm that OsNCED3 acts as a key regulator of ABA biosynthesis and links ABA production with leaf senescence.
Collectively, our observations found that OsNCED3 controls ABA levels in rice to ensure seed dormancy, growth and stomatal aperture, while contributing to responses to severe environmental conditions, which showed OsNCED3 is the key regulator of ABA biosynthesis in rice. Therefore, OsNCED3 is crucial to crop improvement. Moreover, future investigation of OsNCED3 will include determining how OsNCED genes, together with OsNCED3, participate in other physiological functions that we could not observe in the nced3 single mutant. Furthermore, other novel regulatory functions of OsNCED3 could be studied by identifying its interacting proteins.
Author Contributions
YH and ML designed the experiments. YH performed the experiments and wrote the manuscript. YG, YL, and FZ generated the transgenic materials. ZW, FW, and HW provided assistance in the abiotic stress pre-screening experiment. DL, SL, and DM modified the manuscript. ML and LC analyzed the data and edited the manuscript. All authors read and approved the final manuscript draft.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Lijia Qu (College of Life Science, Peking University, China) for providing CRISPR/Cas9 system. This study was supported by the National Natural Science Foundation of China (No. 31171473), the Hunan Province Key Research and Development Plan (No. 2016JC2023), the National Key Research and Development Program of China (No. 2016yFD0101107), the Hunan Provincial Innovation Foundation for Postgraduate (CX2016B171).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00162/full#supplementary-material
References
Agrawal, G. K., Yamazaki, M., Kobayashi, M., Hirochika, R., Miyao, A., and Hirochika, H. (2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 125, 1248–1257. doi: 10.1104/pp.125.3.1248
Bang, S. W., Park, S. H., Jeong, J. S., Kim, Y. S., Jung, H., Ha, S. H., et al. (2013). Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations. Planta 237, 211–224. doi: 10.1007/s00425-012-1764-1
Barrero, J. M., Piqueras, P., Gonzalez-Guzman, M., Serrano, R., Rodriguez, P. L., Ponce, M. R., et al. (2005). A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 56, 2071–2083. doi: 10.1093/jxb/eri206
Bates, L., Waldren, R., and Teare, I. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Burbidge, A., Grieve, T. M., Jackson, A., Thompson, A., McCarty, D. R., and Taylor, I. B. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 17, 427–431. doi: 10.1046/j.1365-313X.1999.00386.x
Cai, S., Jiang, G., Ye, N., Chu, Z., Xu, X., Zhang, J., et al. (2015). A key ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. PLoS ONE 10:e0116646. doi: 10.1371/journal.pone.0116646
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55, 604–619. doi: 10.1093/pcp/pct204
Cheng, W. H., Endo, A., Zhou, L., Penney, J., Chen, H.-C., Arroyo, A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. doi: 10.1105/tpc.006494
Chernys, J. T., and Zeevaart, J. A. (2000). Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 124, 343–353. doi: 10.1104/pp.124.1.343
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. doi: 10.1016/j.plaphy.2011.09.012
Desikan, R., Cheung, M.-K., Bright, J., Henson, D., Hancock, J. T., and Neill, S. J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55, 205–212 doi: 10.1093/jxb/erh033
Endo, A., Sawada, Y., Takahashi, H., Okamoto, M., Ikegami, K., Koiwai, H., et al. (2008). Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 147, 1984–1993. doi: 10.1104/pp.108.116632
Finkelstein, R. (2013). Abscisic Acid synthesis and response. Arabidopsis Book 11:e0166. doi: 10.1199/tab.0166
Frey, A., Boutin, J. P., Sotta, B., Mercier, R., and Marion-Poll, A. (2006). Regulation of carotenoid and ABA accumulation during the development and germination of Nicotiana plumbaginifolia seeds. Planta 224, 622–632. doi: 10.1007/s00425-006-0231-2
Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F., Berger, A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. doi: 10.1111/j.1365-313X.2011.04887.x
Hao, G. P., Zhang, X. H., Wang, Y. Q., Wu, Z. Y., and Huang, C. L. (2009). Nucleotide Variation in the NCED3 Region of Arabidopsis thaliana and its association study with Abscisic acid content under drought stress. J. Integr. Plant Biol. 51, 175–183. doi: 10.1111/j.1744-7909.2008.00786.x
Hauser, F., Waadtl, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Hu, X. L., Jiang, M. Y., Zhang, A. Y., and Lu, J. (2005). Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 223, 57–68. doi: 10.1007/s00425-005-0068-0
Hwang, S. G., Chen, H. C., Huang, W. Y., Chu, Y. C., Shii, C. T., and Cheng, W. H. (2010). Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 178, 12–22. doi: 10.1016/j.plantsci.2009.09.014
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jiang, Y., Qiu, Y., Hu, Y., and Yu, D. (2016). Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front. Plant Sci. 7:145. doi: 10.3389/fpls.2016.00145
Lee, I. C., Hong, S. W., Whang, S. S., Lim, P. O., Nam, H. G., and Koo, J. C. (2011). Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52, 651–662. doi: 10.1093/pcp/pcr026
Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., et al. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. doi: 10.1111/j.1365-313X.2005.02622.x
LeNoble, M. E., Spollen, W. G., and Sharp, R. E. (2004). Maintenance of shoot growth by endogenous ABA: genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 55, 237–245 doi: 10.1093/jxb/erh031
Li, H. Z., and Cheng, Z. H. (2015). Hoagland nutrient solution promotes the growth of cucumber seedlings under light-emitting diode light. Acta Agric. Scand. Sect. B Soil Plant Sci. 65, 74–82. doi: 10.1080/09064710.2014.967285
Li, J., Huang, Y., Tan, H., Yang, X., Tian, L. F., Luan, S., et al. (2015). An endoplasmic reticulum magnesium transporter is essential for pollen development in Arabidopsis. Plant Sci. 231, 212–220. doi: 10.1016/j.plantsci.2014.12.008
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lin, P. C., Hwang, S. G., Endo, A., Okamoto, M., Koshiba, T., and Cheng, W. H. (2007). Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 143, 745–758. doi: 10.1104/pp.106.084103
Lou, D., Wang, H., Liang, G., and Yu, D. (2017). OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 8:993. doi: 10.3389/fpls.2017.00993
Mao, C., Lu, S., Lv, B., Zhang, B., Shen, J., He, J., et al. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174, 1747–1763. doi: 10.1104/pp.17.00542
Martínez-Andújar, C., Ordiz, M. I., Huang, Z., Nonogaki, M., Beachy, R. N., and Nonogaki, H. (2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. U.S.A. 108, 17225–17229. doi: 10.1073/pnas.1112151108
Meng, W., Hsiao, A. S., Gao, C., Jiang, L., and Chye, M. L. (2014). Subcellular localization of rice acyl-CoA-binding proteins (ACBPs) indicates that OsACBP6::GFP is targeted to the peroxisomes. New Phytol. 203, 469–482. doi: 10.1111/nph.12809
Milborrow, B. V. (2001). The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52, 1145–1164. doi: 10.1093/jexbot/52.359.1145
Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Ng, L. M., Melcher, K., Teh, B. T., and Xu, H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin. 35, 567–584. doi: 10.1038/aps.2014.5
Ozfidan, C., Turkan, I., Sekmen, A. H., and Seckin, B. (2012). Abscisic acid-regulated responses of aba2-1 under osmotic stress: the abscisic acid-inducible antioxidant defence system and reactive oxygen species production. Plant Biol. 14, 337–346. doi: 10.1111/j.1438-8677.2011.00496.x
Pedrosa, A. M., Cidade, L. C., Martins, C. P. S., Macedo, A. F., Neves, D. M., Gomes, F. P., et al. (2017). Effect of overexpression of citrus 9-cis-epoxycarotenoid dioxygenase 3 (CsNCED3) on the physiological response to drought stress in transgenic tobacco. Genet. Mol. Res. 16:gmr16019292. doi: 10.4238/gmr16019292
Priya, R., and Siva, R. (2015). Analysis of phylogenetic and functional diverge in plant nine-cis epoxycarotenoid dioxygenase gene family. J. Plant Res. 128, 519–534. doi: 10.1007/s10265-015-0726-7
Qin, X., and Zeevaart, J. A. D. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128, 544–551. doi: 10.1104/pp.010663
Rock, C. D., and Zeevaart, J. A. (1991). The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 88, 7496–7499. doi: 10.1073/pnas.88.17.7496
Schwartz, S. H., Tan, B. C., Gage, D. A., Zeevaart, J. A., and McCarty, D. R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. doi: 10.1126/science.276.5320.1872
Schwartz, S. H., Tan, B. C., McCarty, D. R., Welch, W., and Zeevaart, J. A. D. (2003). Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxygenase in the ABA biosynthetic pathway. Biochim. Biophys. Acta 1619, 9–14. doi: 10.1016/S0304-4165(02)00422-1
Seo, M., Kanno, Y., Frey, A., North, H. M., and Marion-Poll, A. (2016). Dissection of Arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 246, 91–97. doi: 10.1016/j.plantsci.2016.02.013
Sharp, R. E., and LeNoble, M. E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53, 33–37. doi: 10.1093/jexbot/53.366.33
Sharp, R. E., LeNoble, M. E., Else, M. A., Thorne, E. T., and Gherardi, F. (2000). Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: evidence for an interaction with ethylene. J. Exp. Bot. 51, 1575–1584. doi: 10.1093/jexbot/51.350.1575
Shi, L., Guo, M., Ye, N., Liu, Y., Liu, R., Xia, Y., et al. (2015). Reduced ABA accumulation in the root system is caused by ABA exudation in upland rice (Oryza sativa L. var. Gaoshan1) and this enhanced drought adaptation. Plant Cell Physiol. 56, 951–964. doi: 10.1093/pcp/pcv022
Son, S., Chitnis, V. R., Liu, A. H., Gao, F., Nguyen, T. N., and Ayele, B. T. (2016). Abscisic acid metabolic genes of wheat (Triticum aestivum L.): identification and insights into their functionality in seed dormancy and dehydration tolerance. Planta 244, 429–447. doi: 10.1007/s00425-016-2518-2
Song, S. Y., Dai, X. Y., and Zhang, W. H. (2012). A rice F-box gene, OsFbx352, is involved in glucose-delayed seed germination in rice. J. Exp. Bot. 63, 5559–5568. doi: 10.1093/jxb/ers206
Tan, B.-C., Joseph, L. M., Deng, W.-T., Liu, L., Li, Q.-B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56. doi: 10.1046/j.1365-313X.2003.01786.x
Tan, B. C., Schwartz, S. H., Zeevaart, J. A., and McCarty, D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94, 12235–12240. doi: 10.1073/pnas.94.22.12235
Tardieu, F., and Davies, W. J. (1992). Stomatal response to abscisic Acid is a function of current plant water status. Plant Physiol. 98, 540–545. doi: 10.1104/pp.98.2.540
Taylor, I. B., Burbidge, A., and Thompson, A. J. (2000). Control of abscisic acid synthesis. J. Exp. Bot. 51, 1563–1574. doi: 10.1093/jexbot/51.350.1563
Teng, K., Li, J., Liu, L., Han, Y., Du, Y., Zhang, J., et al. (2014). Exogenous ABA induces drought tolerance in upland rice: the role of chloroplast and ABA biosynthesis-related gene expression on photosystem II during PEG stress. Acta Physiol. Plantarum 36, 2219–2227. doi: 10.1007/s11738-014-1599-4
Toh, S., Imamura, A., Okamoto, M., Kamiya, Y., Nambara, E., and Kawakami, N. (2007). Unequally redundant functions of NCED9 and NCED5 genes in thermoinhibition of Arabidopsis seed germination. Plant Cell Physiol. 48, S27–S27
Tung, S. A., Smeeton, R., White, C. A., Black, C. R., Taylor, I. B., Hilton, H. W., et al. (2008). Over-expression of LeNCED1 in tomato (Solanum lycopersicum L.) with the rbcS3C promoter allows recovery of lines that accumulate very high levels of abscisic acid and exhibit severe phenotypes. Plant Cell Environ. 31, 968–981. doi: 10.1111/j.1365-3040.2008.01812.x
Voetberg, G. S., and Sharp, R. E. (1991). Growth of the maize primary root at low water potentials: III. Role of increased proline deposition in osmotic adjustment. Plant Physiol. 96, 1125–1130. doi: 10.1104/pp.96.4.1125
Wang, P. T., and Song, C. P. (2008). Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 178, 703–718. doi: 10.1111/j.1469-8137.2008.02431.x
Xia, H., Wu, S., and Ma, F. (2014). Cloning and expression of two 9-cis-epoxycarotenoid dioxygenase genes during fruit development and under stress conditions from Malus. Mol. Biol. Rep. 41, 6795–6802. doi: 10.1007/s11033-014-3565-z
Xu, P., and Cai, W. (2017). Functional characterization of the BnNCED3 gene in Brassica napus. Plant Sci. 256, 16–24. doi: 10.1016/j.plantsci.2016.11.012
Xu, Z.-Y., Kim, D. H., and Hwang, I. (2013). ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 32, 807–813. doi: 10.1007/s00299-013-1396-3
Yang, J., and Guo, Z. (2007). Cloning of a 9-cis-epoxycarotenoid dioxygenase gene (SgNCED1) from Stylosanthes guianensis and its expression in response to abiotic stresses. Plant Cell Rep. 26, 1383–1390. doi: 10.1007/s00299-007-0325-8
Yao, Y., Liu, X., Li, Z., Ma, X., Rennenberg, H., Wang, X., et al. (2013). Drought-induced H2O2 accumulation in subsidiary cells is involved in regulatory signaling of stomatal closure in maize leaves. Planta 238, 217–227. doi: 10.1007/s00425-013-1886-0
Ye, N., Jia, L., and Zhang, J. (2012). ABA signal in rice under stress conditions. Rice 5:1. doi: 10.1186/1939-8433-5-1
Ye, N., Zhu, G., Liu, Y., Li, Y., and Zhang, J. (2011). ABA controls H2O2 accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol. 52, 689–698. doi: 10.1093/pcp/pcr028
Zhang, D. P., Zhou, Y., Yin, J. F., Yan, X. J., Lin, S., Xu, W. F., et al. (2015). Rice G-protein subunits qPE9-1and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J. Exp. Bot. 66, 6371–6384. doi: 10.1093/jxb/erv350
Zhang, L., Xiao, S. S., Li, W. Q., Feng, W., Li, J., Wu, Z. D., et al. (2011). Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J. Exp. Bot. 62, 4229–4238. doi: 10.1093/jxb/err131
Zhu, C., Kauder, F., Röemer, S., and Sandmann, G. (2007). Cloning of two individual cDNAS encoding 9-cis-epoxycarotenoid dioxygenase from Gentiana lutea, their tissue-specific expression and physiological effect in transgenic tobacco. J. Plant Physiol. 164, 195–204. doi: 10.1016/j.jplph.2006.02.010
Keywords: OsNCED3, abscisic acid (ABA), CRISPR/Cas9 system, seed germination, growth, abiotic stress, leaf senescence
Citation: Huang Y, Guo Y, Liu Y, Zhang F, Wang Z, Wang H, Wang F, Li D, Mao D, Luan S, Liang M and Chen L (2018) 9-cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 9:162. doi: 10.3389/fpls.2018.00162
Received: 07 November 2017; Accepted: 29 January 2018;
Published: 06 March 2018.
Edited by:
Paul E. Verslues, Academia Sinica, TaiwanReviewed by:
Tyler Dowd, Donald Danforth Plant Science Center, United StatesBhaskara Govinal Badiger, University of Texas at Austin, United States
Copyright © 2018 Huang, Guo, Liu, Zhang, Wang, Wang, Wang, Li, Mao, Luan, Liang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manzhong Liang, 1962liang@163.com
Liangbi Chen, chenliangbi@126.com
 Yuan Huang
Yuan Huang Manzhong Liang
Manzhong Liang