- Scion, Rotorua, New Zealand
New Zealand (NZ) is a small country with an export-led economy with above 90% of primary production exported. Plant-based primary commodities derived from the pastoral, horticultural and forestry sectors account for around half of the export earnings. Productivity is characterized by a history of innovation and the early adoption of advanced technologies. Gene editing has the potential to revolutionize breeding programmes, particularly in NZ. Here, perennials such as tree crops and forestry species are key components of the primary production value chain but are challenging for conventional breeding and only recently domesticated. Uncertainty over the global regulatory status of gene editing products is a barrier to invest in and apply editing techniques in plant breeding. NZs major trading partners including Europe, Asia and Australia are currently evaluating the regulatory status of these technologies and have not made definitive decisions. NZ is one of the few countries where the regulatory status of gene editing has been clarified. In 2014, the NZ Environmental Protection Authority ruled that plants produced via gene editing methods, where no foreign DNA remained in the edited plant, would not be regulated as GMOs. However, following a challenge in the High Court, this decision was overturned such that NZ currently controls all products of gene editing as GMOs. Here, we illustrate the potential benefits of integrating gene editing into plant breeding programmes using targets and traits with application in NZ. The regulatory process which led to gene editing's current GMO classification in NZ is described and the importance of globally harmonized regulations, particularly to small export-driven nations is discussed.
Introduction
Primary exports are critical to New Zealand's (NZ's) economy providing both employment and export revenue. In 2017, this totalled NZ$38 billion of which the dairy industry contributed NZ$ 14.6 billion, red meat and wool NZ$8.4 billion, forestry NZ$5.5 billion and horticulture NZ$5.1 billion (Ministry for Primary Industries, 2018b). New Zealand's pasture-based dairy industry is the world's largest dairy exporter and accounts for a third of the world's dairy trade (Chobtang et al., 2017a). Sheep and beef make up the majority of animal-based exports but venison and wool are significant contributors. The NZ sheep and beef sector exports close to 90% of its production. Forestry, based around exotic plantation forests (primarily radiata pine and Douglas-fir), covers 1.751 million hectares—approximately 7% of NZ's land area (Ministry for Primary Industries, 2018a). The horticultural sector is predominately fruit based and led by kiwifruit, of which 95% of production is exported, wine, apple and pear are also exported in significant volumes. The main destinations for primary exports are China (NZ$9.1 billion), Australia (NZ$4.3 billion), and the US (NZ$4.0 billion), with Japan, South Korea and Europe also being significant markets.
Nations with small domestic markets like NZ face pressure to continuously adjust and innovate in order to maintain global competitiveness (Vitalis, 2007). To support this, NZ has a long history of implementation of agritech innovation (Easton, 1997; Vitalis, 2007; Hedley, 2015) including the use of genetic technologies (Harris et al., 2009; Kumar et al., 2012). In order to maintain NZ's position whilst providing sustainable solutions to the challenges of global food security and climate change a step change in productivity beyond that which has been possible through conventional breeding will be required (Williams et al., 2007). Solutions are also urgently required for the increased threat from pests and diseases. In the last decade the kiwifruit and forestry industry have suffered considerable losses from emerging diseases (Vanneste, 2012; Scott and Williams, 2014). Myrtle rust, which has caused worldwide damage to both agricultural and native ecosystems, arrived in NZ in 2017 (Office of the Minister of Conservation, 2017). Biotechnology-based improvements have the potential to be an important tool in delivering this. The unprecedented uptake of genetically modified (GM) crops over the last 20 years, such that 189.8 million hectares of GM crops were planted in 24 countries in 2017 (ISAAA, 2017) is testimony to this. GM crops are now cultivated on more than 10% of the worlds farmland and comprise 80% of global cotton and 77% of soybean plantings (ISAAA, 2017; Taheri et al., 2017).
Currently no GM crops are grown in NZ. The globally traded cash crops (corn, soybean, canola and cotton) that make up the majority of current GM plantings are not widely grown and do not provide a compelling value proposition for NZ. In contrast, NZ aims to supply high value innovative products that are not cultivated on a global large scale e.g., kiwifruit and radiata pine. The time and cost of developing and gaining regulatory approval for GM versions of these for the NZ market is prohibitive. The lack of relevant GM crops has meant that there has not been recent nationwide debate on the merits of these technologies in NZ (Bryan and Roberts, 2015).
Over the last decade genome editing methods based on Zinc finger nucleases (Urnov et al., 2010), TALENs (Chen and Gao, 2013), CRISPR/Cas (Doudna and Charpentier, 2014) systems have rapidly revolutionized both basic and applied biology. The wide-ranging applications of this technology have been extensively reviewed elsewhere (Voytas, 2013; Carroll, 2014; Wang et al., 2016a; Brooks and Gaj, 2018). In this review, we will focus on the use of gene editing to carry out targeted mutagenesis on plant species where no DNA template is used. We believe this technology has the ability to encourage a paradigm shift in the incorporation of biotechnology into NZ plant breeding programmes. Particularly if, as seems likely, it is ultimately regulated in a less burdensome way than GM technology. Here, we give examples of the traits that could be modified to give NZ relevant outcomes, describe the current regulatory landscape, and discuss the implications of this on the future innovation in NZ plant-based primary industries.
Potential Applications of Gene Editing in New Zealand
Gene editing offers the potential to produce a step change in NZ primary industry productivity, biosecurity and speed of innovation. This is particularly the case for perennial crops with slow or complex breeding cycles that are a feature of NZ's plant-based exports. Although gene editing has already been demonstrated for a number of NZ relevant crops (Table 1), it is still to be implemented for a number of important species particularly conifer forestry species. This review focuses on plant-based applications, however, uses in animal breeding (Wei et al., 2018) and control of introduced pests via gene drive technology (Dearden et al., 2018) are also in development. Below, as examples, we describe possible scenarios where plant-based gene editing could have an impact on primary production and innovation.
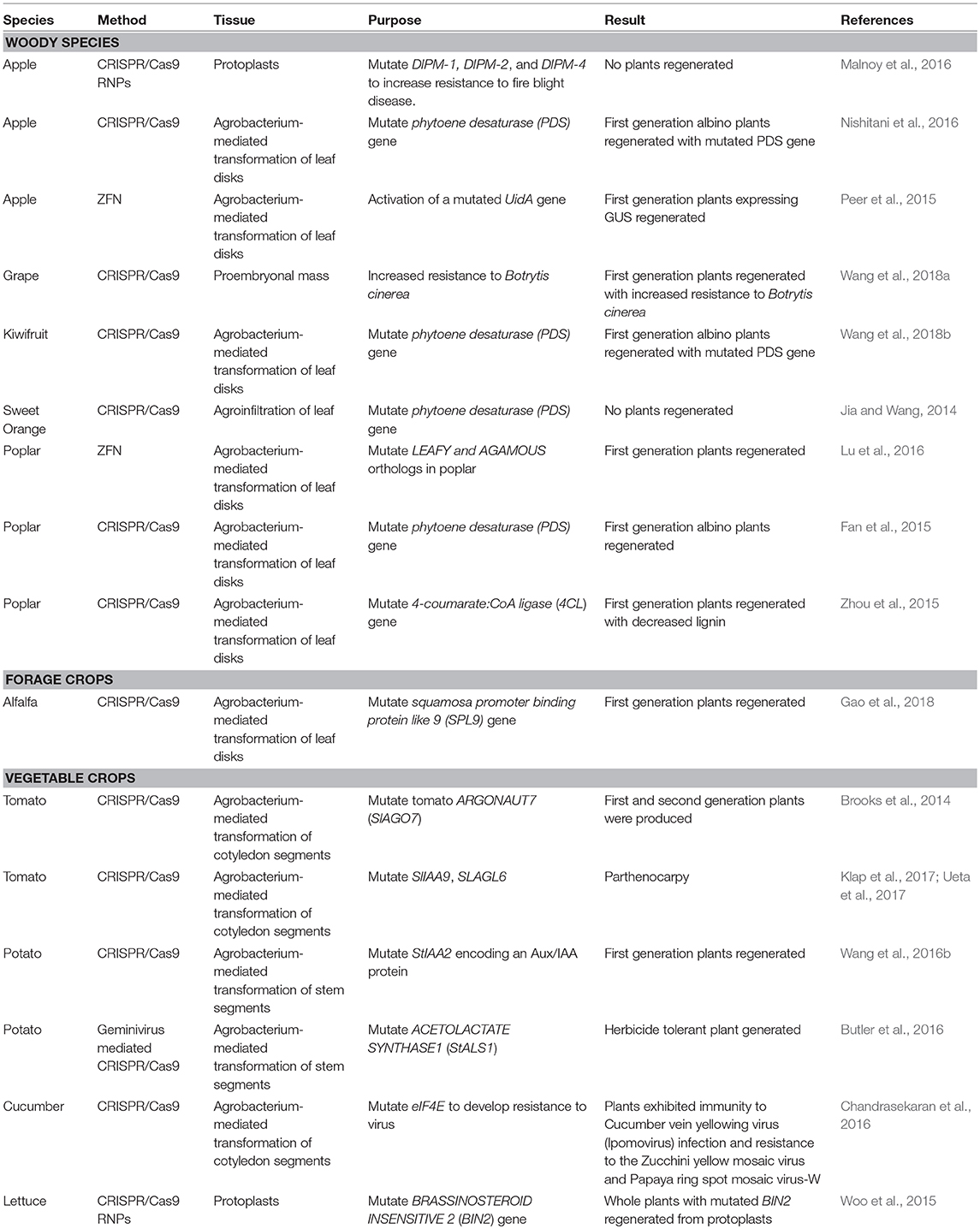
Table 1. Examples of species relevant to New Zealand's plant-based primary industries that have been modified using genome editing technologies.
Control of Invasive Conifers by Manipulation of Reproduction
NZ faces serious ecological, economic and cultural challenges from invasive tree species that have “escaped” by seed dispersal from planted forests and shelter belts (Richardson and Rejmánek, 2004). Several exotic conifer species that have become established outside plantations now occupy ~1.8 million ha of land, and are expanding by 6% annually (Froude, 2011). The government has declared these to be the most significant weed problem facing NZ (The New Zealand Government, 2016a) with control of the existing population costing an estimated NZ$15 million each year. The social and economic costs of these escapes is challenging the ability of forest owners to carry out new plantings with commercially advantageous, but potentially invasive species such as Douglas-fir. The capability to generate trees that are unable to reproduce would allow control programs to focus on the existing populations and give freedom to operate for new plantings. Prevention of cone development is also predicted to increase growth and wood development by the redirection of energy and nutrients toward vegetative growth (Santos-del-Blanco and Climent, 2014).
Gene editing provides an attractive approach to prevent the generation of new escapees via targeted mutagenesis of genes essential for normal sexual reproduction. Genes involved in the transition from the juvenile to reproductive growth phase, cone initiation or development, and pollen formation and development are potential targets (Strauss et al., 1995). If transgene-free edited trees are required, DNA-free delivery methods would be necessary because the long breeding cycles of conifers would prevent timely segregation of transgenes from edited genes.
Rapid Breeding in Apple
Breeding of new apple varieties is a slow process limited by a long-lasting juvenile stage taking more than two decades to bring a new variety into the market (Flachowsky et al., 2009). Shortening the juvenile stage has been the subject of intensive research and is a major objective in breeding (Meilan, 1997). Early flowering has been demonstrated in apple through the overexpression of beech MADS4 and Arabidopsis FT gene (Flachowsky et al., 2007; Yamagishi et al., 2011). This technology has been used to rapidly breed fire blight resistance into apple within 7 years (Schlathölter et al., 2018). A similar result has been obtained using antisense-based silencing of MdTFL1 expression (Kotoda et al., 2006). Gene editing could be used to knock out the expression of MdTFL1 to reproduce this early flowering phenotype. This would allow rapid breeding of new cultivars through several cycles after which the edited gene could be crossed out to restore the non-engineered flowering phenotype without any trace of the modification.
Improved Pasture Quality
The dairy, meat and wool industries in NZ draw a significant market advantage from the predominantly pasture-based feed. Limiting environmental impacts whilst meeting the increase in global demand for dairy products requires improvements in pasture productivity (Chobtang et al., 2017b). Forage pastures generally consist of ryegrass, alfalfa and clover. Of these, annual and perennial ryegrass are most common. Gene editing provides tools to improve productivity and reduce disease either through the direct manipulation of forage crops or via manipulation of endophytes. The incorporation of herbicide tolerance (Butler et al., 2016) and easier digestibility (Li et al., 2018) have both been successfully introduced into plants by gene editing and research to increase energy values is underway. These are likely to offer routes to both increased productivity and a reduced environmental footprint.
Forage grasses like ryegrass are usually infected with symbiotic fungal endophytes (Latch et al., 1984) which produce secondary metabolites that protect the plant from invertebrate pests (Mortimer and Di Menna, 1983), give higher growth rates, tolerance to abiotic stress (West and Gwinn, 1993), and produce more dry matter than non-infected plants (Popay et al., 1999). These benefits can be compromised by the production of high levels of indole-diterpenes and alkaloids that have negative impacts on livestock e.g., ryegrass staggers in sheep (Fletcher and Harvey, 1981; Thom et al., 2007). To minimize the toxicity of these symbionts, strains of endophytes were selected that produced low levels of these alkaloids and indole-diterpenes (Davies et al., 1993). Molecular analysis revealed these lower levels were due to deletions within the coding sequence of genes in the biosynthetic pathway (Young et al., 2009). Gene editing will allow the modification of biosynthetic pathways to decrease or eliminate toxins and increase the production of desirable metabolites without the need to screen for extremely rare natural variants.
Regulation of Gene Editing in New Zealand
The global social and regulatory landscape surrounding GM crops remains complex with many different regulatory systems in place (Wolt et al., 2016; Davison and Ammann, 2017). The primary difference being whether a process or product driven framework is used (Ishii and Araki, 2017). As yet there is not a global consensus on the regulation of gene editing which was developed after current regulatory frameworks were put in place. Several nations, including the USA, Canada and Argentina, have decided that gene editing technologies where the final plant does not contain introduced DNA will not be regulated (Whelan and Lema, 2015; Ishii and Araki, 2017; Waltz, 2018). In contrast the European Union recently decided that all gene editing technologies will be regulated in the same way as conventional GM organisms (Callaway, 2018; Kupferschmidt, 2018). Others, including the two main destinations for NZ's primary exports, China and Australia, are yet to decide on their regulatory approach.
New Zealand regulates GM organisms using a stringent process driven regulatory framework—the Hazardous Substances and New Organisms (HSNO) Act 1996. The Act defines a GMO very broadly as any organism where the genes or genetic material have been modified by in vitro techniques (Table 2a). A number of technologies that were in use at the time the Act was passed are captured by this broad definition e.g., somaclonal variation, cell fusion, and chemical and physical mutagenesis. To counter this, a number of technologies that meet the definition of generating a GMO are excluded from being regulated by the HSNO (Organisms Not Genetically Modified) Regulations 1998 (Table 2b).
Application to Determine Status of Gene Editing
The HSNO Act, under section 26, provides a mechanism for an applicant to ask for a determination by the Environmental Protection Authority (EPA) as to whether, an organism is regulated as a GM in NZ (Kershen, 2015). In 2012, Scion, a forestry-focused Crown Research Institute, used this procedure to seek a determination on how gene edited organisms would be regulated. The HSNO definition (Table 2a) includes a clause specifying that genetic modifications “inherited or otherwise derived, through any number of replications” would be classed as GMOs. Scion's application, which was submitted before CRISPR/Cas9 technology was developed, thus sought to determine “whether the use of custom Zinc Finger Nucleases and custom Transcription Activator-Like Effectors results in organisms classed as genetically modified organisms” when the editing complex was delivered without the use of a transgene to carry the editing machinery.
Scion's application argued that gene editing technologies that did not include the insertion of a transgene into host genome were similar in process and outcome to chemical mutagenesis. As such they should be included within the HSNO regulations exception of “chemical or radiation treatments that cause changes in chromosome number or cause chromosome rearrangements” (Table 2b). Scion noted that the list of techniques that were excluded from regulation was preceded by the word included (underlined for emphasis in Table 2b) suggesting that these were example techniques and not a closed list.
EPA Decision
In their decision of April 2013, the EPA concluded that the non-transgenic gene editing approach proposed by Scion had similarities to both chemical mutagenesis and genetic manipulation. However, because the changes involved the use of a chemical agent (in this case, a protein) without the introduction of foreign DNA it is more similar to chemical mutagenesis (Environmental Protection Authority, 2013). The EPA further stated that the Regulations (Table 2b) exclude products of chemical mutagenesis from being regulated as GMOs under the Act and that the proposed modifications were sufficiently similar to those listed in the Regulations and should also be excluded, and organisms arising from them should not be considered GMOs.
High Court Challenge
The EPA decision was appealed by the Sustainability Council of New Zealand in the High Court and the case was heard in November 2013. The key consideration of the judgement, issued on the 20th May 2014, was “whether the specific techniques (listed in Table 2b) are a closed list of techniques that are exempted, or whether they describe a category of the kind of techniques that are excepted (so that other techniques which are sufficiently similar to those techniques are also exempted)” (The High Court of New Zealand, 2014). The Court concluded that the list of techniques listed in the HSNO (Organisms Not Genetically Modified) Regulations 1998 (Table 2b) are a closed list and that adding to the exceptions list is a political decision and not an administrative decision (Kershen, 2015). On this basis the EPA's original decision was quashed and all gene editing is currently regulated as a GM procedure in NZ.
Implications of the Decision
In the court ruling the judge pointed out that the regulations are not well drafted, brackets are in the wrong place and the grammar poor. This reinforced her interpretation that the unorthodox use of the word “including” before start of the list of techniques that do not produce GMOs (Table 2b) does not constitute a list of examples but rather a closed list. She also highlighted that the regulations exempted only “chemical or radiation treatments that cause changes in chromosome number or cause chromosome rearrangements” from regulation as GMOs. Some long-standing in vitro chemical treatments do not have these effects, but are caught by this definition. Thus, techniques such as EMS mutagenesis that cause point mutations rather than changes in chromosome number or chromosome rearrangements are regarded technically as GMOs.
In response to these inconsistencies the government held a review of the not genetically modified regulations. The review, which included a public consultation process, resulted in changes intended to maintain the intent of the 1998 regulations and address the drafting errors present in the original regulations. The wording was changed such that mutagenesis techniques that were in use before 1998 were not regulated whilst those developed later are regulated as GMOs. This was done by simply excluding from regulation “mutagenesis that uses chemical or radiation treatments that were in use on or before 29 July 1998” (Table 2c) (The New Zealand Government, 2016b). Mutagenesis techniques developed later, including gene editing, however similar they are to the original excluded techniques are regulated as GMOs.
Future Outlook
Gene editing continues to rapidly evolve with developments such as new enzyme capabilities (Yin et al., 2018), base editing (Komor et al., 2016) and simultaneous multi-target approaches, (Svitashev et al., 2015; Chilcoat et al., 2017; Shen et al., 2017) increasing the scope and applicability of the technology. The recent demonstration of rapid de novo gene editing-based domestication of wild type relatives of domestic crops without the need for a long breeding programme (Zsögön et al., 2017) has particular applicability in NZ. Particular examples are kiwifruit and radiata pine which are relatively undomesticated and/or where a large number of wildtype genotypes are available (Ferguson, 2007) but require the introduction of essential commercial traits such as longer post-harvest storage and shelf life.
Recent decisions in USA (Waltz, 2018) and the UK (Rogowsky and Wilhelm, 2018) indicate that crops produced using gene editing-based targeted mutagenesis will be able to go to market without going through a time-consuming and burdensome regulatory process required for GMO crops. This regulatory approach will drastically reduce the time to market and compliance costs for gene edited crops. The recent US Department of Agriculture (USDA) approval of Camelina sativa edited for enhanced omega-3 oil was completed in 2 years at a much lower cost than the estimated US$30-50 million and 6 years plus that would have been required to fulfill the full USDA process (Waltz, 2018).
In contrast, NZ has adopted a wait-and-see-approach with regard to the regulation of gene editing. The government indicating that a cautious approach is appropriate because as an exporter of billions of dollars of food products we need to be mindful of market perceptions as well as the science (The New Zealand Government, 2016b). It should be noted that the three largest importers of NZ primary products, China, Australia and USA all currently grow GM crops and Australia and China seem likely to follow the lead of USA in not regulating gene edited crops. The current NZ approach prevents rapid implementation of non-transgenic gene editing and also places the extremely high regulatory compliance costs associated with GM research on developers of such technology.
For NZ to maintain its current global competiveness it is essential that industry is able to continue to implement innovative solutions. For this to happen with gene editing, it will be necessary for the government to be proactive in ensuring NZ is in step with global competitors and that innovation is not stifled by the current outdated regulations. Despite the opinion released in January, by the advocate-general of the European Court of Justice, that gene edited crops that did not contain foreign DNA could be exempted from the GMO regulations, the EU has recently decided to adopt a similar regulatory approach to that of NZ. All gene edited crops will be subject to the same stringent regulations as conventional genetically engineered organisms (Callaway, 2018). This makes a global consensus on regulation of gene editing impossible in the immediate future. Although it is too early to judge the long-term impacts of this decision on the global uptake of gene editing or the regulatory approach that will be taken by currently undecided nations, the existence of different regulatory systems will undoubtedly create many challenges, particularly for those nations with strong trading links with the EU.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by Scion's Strategic Science Investment Funding (SSIF) from the Science and Innovation Group, Ministry of Innovation, Business and Science.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Brooks, A. K., and Gaj, T. (2018). Innovations in CRISPR technology. Curr. Opin. Biotechnol. 52, 95–101. doi: 10.1016/j.copbio.2018.03.007
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Bryan, G., and Roberts, N. (2015). Superior Forages Could Result From GM Technology. Technical Series Online, 1–5.
Butler, N. M., Baltes, N. J., Voytas, D. F., and Douches, D. S. (2016). Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 7:1045. doi: 10.3389/fpls.2016.01045
Callaway, E. (2018). CRISPR plants now subject to tough GM laws in European Union. Nature 560:16. doi: 10.1038/d41586-018-05814-6
Carroll, D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. doi: 10.1146/annurev-biochem-060713-035418
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chen, K., and Gao, C. (2013). TALENs: customizable molecular DNA scissors for genome engineering of plants. J. Genet. Genomics 40, 271–279. doi: 10.1016/j.jgg.2013.03.009
Chilcoat, D., Liu, Z.-B., and Sander, J. (2017). “Use of CRISPR/Cas9 for crop improvement in maize and soybean,” in Progress in Molecular Biology and Translational Science, eds D. P. Weeks, and B. Yang (San Diego, CA: Elsevier), 27–46.
Chobtang, J., McLaren, S. J., Ledgard, S. F., and Donaghy, D. J. (2017a). Consequential life cycle assessment of pasture-based milk production: a case study in the Waikato region, New Zealand. J. Ind. Ecol. 21, 1139–1152. doi: 10.1111/jiec.12484
Chobtang, J., McLaren, S. J., Ledgard, S. F., and Donaghy, D. J. (2017b). Environmental trade-offs associated with intensification methods in a pasture-based dairy system using prospective attributional life cycle assessment. J. Clean. Prod. 143, 1302–1312. doi: 10.1016/j.jclepro.2016.11.134
Davies, E., Lane, G., Latch, G., Tapper, B., Garthwaite, I., Towers, N., et al. (1993). “Alkaloid concentrations in field-grown synthetic perennial ryegrass endophyte associations,” in Proceedings of the Second International Symposium on Acremonium/Grass Interactions (Palmerston North), 72–76.
Davison, J., and Ammann, K. (2017). New GMO regulations for old: determining a new future for EU crop biotechnology. GM Crops Food 8, 13–34. doi: 10.1080/21645698.2017.1289305
Dearden, P. K., Gemmell, N. J., Mercier, O. R., Lester, P. J., Scott, M. J., Newcomb, R. D., et al. (2018). The potential for the use of gene drives for pest control in New Zealand: a perspective. J. R. Soc. NZ. 48, 225–244. doi: 10.1080/03036758.2017.1385030
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Environmental Protection Authority (2013). Decision. Available online at: https://www.epa.govt.nz/assets/FileAPI/hsno-ar/APP201381/APP201381-APP201381-Decision.pdf (Accessed 3 July 2018).
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Ferguson, A. R. (2007). The need for characterisation and evaluation of germplasm: kiwifruit as an example. Euphytica 154, 371–382. doi: 10.1007/s10681-006-9188-2
Flachowsky, H., Hanke, M. V., Peil, A., Strauss, S., and Fladung, M. (2009). A review on transgenic approaches to accelerate breeding of woody plants. Plant Breeding 128, 217–226. doi: 10.1111/j.1439-0523.2008.01591.x
Flachowsky, H., Peil, A., Sopanen, T., Elo, A., and Hanke, V. (2007). Overexpression of BpMADS4 from silver birch (Betula pendula Roth.) induces early-flowering in apple (Malus × domestica Borkh.). Plant Breeding 126, 137–145. doi: 10.1111/j.1439-0523.2007.01344.x
Fletcher, L., and Harvey, I. (1981). An association of a Lolium endophyte with ryegrass staggers. NZ. Vet. J. 29, 185–186. doi: 10.1080/00480169.1981.34839
Froude, V. A. (2011). Wilding Conifers in New Zealand: Beyond the Status Report. Report prepared for the Ministry of Agriculture and Forestry, Pacific Eco-Logic, Bay of Islands 44.
Gao, R., Feyissa, B. A., Croft, M., and Hannoufa, A. (2018). Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta 247, 1043–1050. doi: 10.1007/s00425-018-2866-1
Harris, B., Johnson, D., and Spelman, R. (2009). Genomic Selection in New Zealand and the Implications for National Genetic Evaluation. ICAR Technical Series, 325–330.
Hedley, C. (2015). The role of precision agriculture for improved nutrient management on farms. J. Sci. Food Agric. 95, 12–19. doi: 10.1002/jsfa.6734
ISAAA (2017). Global Status of Commercialized Biotech/GM Crops in 2017: Biotech Crop Adoption Surges as Economic Benefits Accumulate in 22 years Ithaca, NY: ISAAA.
Ishii, T., and Araki, M. (2017). A future scenario of the global regulatory landscape regarding genome-edited crops. GM Crops Food 8, 44–56. doi: 10.1080/21645698.2016.1261787
Jia, H., and Wang, N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806. doi: 10.1371/journal.pone.0093806
Kershen, D. L. (2015). Sustainability Council of New Zealand Trust v. The Environmental Protection Authority: Gene editing technologies and the law. GM Crops Food 6, 216–222. doi: 10.1080/21645698.2015.1122859
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2017). Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi: 10.1111/pbi.12662
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nat 533, 420–424. doi: 10.1038/nature17946
Kotoda, N., Iwanami, H., Takahashi, S., and Abe, K. (2006). Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J. Am. Soc. Hortic. Sci. 131, 74–81.
Kumar, S., Bink, M. C., Volz, R. K., Bus, V. G., and Chagn,é, D. (2012). Towards genomic selection in apple (Malus × domestica Borkh.) breeding programmes: prospects, challenges and strategies. Tree Genet. Genomes 8, 1–14. doi: 10.1007/s11295-011-0425-z
Kupferschmidt, K. (2018). EU Verdict on CRISPR Crops Dismays Scientists. Washington, DC: American Association for the Advancement of Science.
Latch, G., Christensen, M., and Samuels, G. (1984). Five endophytes of Lolium and Festuca in New Zealand. Mycotaxon 20, 535–550.
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S., Zhang, C., et al. (2018). Editing of an alpha-kafirin gene family increases digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438.doi: 10.1104/pp.18.00200
Lu, H., Klocko, A. L., Dow, M., Ma, C., Amarasinghe, V., and Strauss, S. H. (2016). Low frequency of zinc-finger nuclease-induced mutagenesis in Populus. Mol. Breed. 36:121. doi: 10.1007/s11032-016-0546-z
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Meilan, R. (1997). Floral induction in woody angiosperms. New Forests 14, 179–202. doi: 10.1023/A:1006560603966
Ministry for Primary Industries (2018a). Primary Production, ed L. Policy. Available online at: www.mpi.govt.nz
Ministry for Primary Industries (2018b). Situation and Outlook for Primary Industries (SOPI) March 2018, ed E. I. Unit. Available online at: www.mpi.govt.nz
Mortimer, P., and Di Menna, M. E. (1983). Ryegrass staggers: further substantiation of a Lolium endophyte aetiology and the discovery of weevil resistance of ryegrass pastures infected with Lolium endophyte. Proc. NZ. Grassland Association 44, 240–243.
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 31481. doi: 10.1038/srep31481
Office of the Minister of Conservation (2017). “Myrtle rust found in New Zealand,” in The Fungal Disease Myrtle Rust Has Been Found on Mainland New Zealand and a Response is Underway, ed G. Cavill (Wellington: Offices of the Minister of Conservation), 1–2.
Peer, R., Rivlin, G., Golobovitch, S., Lapidot, M., Gal-On, A., Vainstein, A., et al. (2015). Targeted mutagenesis using zinc-finger nucleases in perennial fruit trees. Planta 241, 941–951. doi: 10.1007/s00425-014-2224-x
Popay, A., Hume, D., Baltus, J., Latch, G., Tapper, B., Lyons, T., et al. (1999). Field performance of perennial ryegrass (Lolium perenne) infected with toxin-free fungal endophytes (Neotyphodium spp.) Ryegrass endophyte: an essential New Zealand symbiosis. Grassland Res. Pract. Ser. 7, 113–122.
Richardson, D. M., and Rejmánek, M. (2004). Conifers as invasive aliens: a global survey and predictive framework. Divers. Distrib. 10, 321–331. doi: 10.1111/j.1366-9516.2004.00096.x
Rogowsky, P., and Wilhelm, R. (2018). Welcoming the Advice on Genome-Edited Camelina Plants by the British Advisory Committee on Releases to the Environment (ACRE). Brussels: European Plant Science Organisation.
Santos-del-Blanco, L., and Climent, J. (2014). Costs of female reproduction in a conifer tree: a whole-tree level assessment. J. Ecol. 102, 1310–1317. doi: 10.1111/1365-2745.12283
Schlathölter, I., Jänsch, M., Flachowsky, H., Broggini, G. A. L., Hanke, M.-V., and Patocchi, A. (2018). Generation of advanced fire blight-resistant apple (Malus × domestica) selections of the fifth generation within 7 years of applying the early flowering approach. Planta 247, 1475–1488. doi: 10.1007/s00425-018-2876-z
Shen, L., Hua, Y., Fu, Y., Li, J., Liu, Q., Jiao, X., et al. (2017). Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci. China Life Sci. 60, 506–515. doi: 10.1007/s11427-017-9008-8
Strauss, S. H., Rottmann, W. H., Brunner, A. M., and Sheppard, L. A. (1995). Genetic engineering of reproductive sterility in forest trees. Mol. Breed. 1, 5–26. doi: 10.1007/BF01682086
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene Insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Taheri, F., Azadi, H., and D'Haese, M. (2017). A world without hunger: organic or GM crops? Sustainability 9:580. doi: 10.3390/su9040580
The High Court of New Zealand (2014). The Sustainability Council of New Zealand Trust v The Environmental Protection Authority. Wellington: The High Court of New Zealand.
The New Zealand Government (2016a). $16m New Funding to Tackle Wilding Conifers, ed Releases. Available online at: https://www.beehive.govt.nz/release/16m-new-funding-tackle-wilding-conifers
The New Zealand Government (2016b). GMO Regulations Clarified. Available online at: https://www.beehive.govt.nz, https://www.beehive.govt.nz/release/gmo-regulations-clarified (Accessed July 4, 2018).
Thom, E., Waugh, C., Minnee, E., and Waghorn, G. (2007). “A new generation ryegrass endophyte–the first results from dairy cows fed AR37,” in Proceedings of the 6th International Symposium on Fungal Endophytes of Grasses (Christchurch: Grassland Research and Practice Series), 293–296.
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507. doi: 10.1038/s41598-017-00501-4
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., and Gregory, P. D. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. doi: 10.1038/nrg2842
Vanneste, J. (2012). Pseudomonas syringae pv. actinidiae (Psa): a threat to the New Zealand and global kiwifruit industry. New Zeal. J. Crop Hort. Sci. 40, 265–267. doi: 10.1080/01140671.2012.736084
Vitalis, V. (2007). Trade, Innovation and Growth: The Case of the New Zealand Agriculture Sector. Paris: Citeseer.
Voytas, D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. doi: 10.1146/annurev-arplant-042811-105552
Waltz, E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36, 6–7. doi: 10.1038/nbt0118-6b
Wang, H., La Russa, M., and Qi, L. S. (2016a). CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wang, X., Niu, Y., Zhou, J., Yu, H., Kou, Q., Lei, A., et al. (2016b). Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 6:32271. doi: 10.1038/srep32271
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., and Li, Z. (2018a). eCRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Wang, Z., Wang, S., Li, D., Zhang, Q., Li, L., Zhong, C., et al. (2018b). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol J. 16, 1424–1433. doi: 10.1111/pbi.12884
Wei, J., Wagner, S., Maclean, P., Brophy, B., Cole, S., Smolenski, G., et al. (2018). Cattle with a precise, zygote-mediated deletion safely eliminate the major milk allergen beta-lactoglobulin. Sci. Rep. 8:7661. doi: 10.1038/s41598-018-25654-8
West, C., and Gwinn, K. (1993). “Role of acremonium in drought, pest, and disease tolerances of grasses,” in Proceedings of the Second International Symposium on Acremonium/Grass Interactions (Palmerston North).
Whelan, A. I., and Lema, M. A. (2015). Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM Crops Food 6, 253–265. doi: 10.1080/21645698.2015.1114698
Williams, W., Easton, H., and Jones, C. (2007). Future options and targets for pasture plant breeding in New Zealand. New Zeal. J. Agr. Res. 50, 223–248. doi: 10.1080/00288230709510292
Wolt, J. D., Wang, K., and Yang, B. (2016). The regulatory status of genome-edited crops. Plant Biotechnol. J. 14, 510–518. doi: 10.1111/pbi.12444
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Yamagishi, N., Sasaki, S., Yamagata, K., Komori, S., Nagase, M., Wada, M., et al. (2011). Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the apple latent spherical virus vector. Plant Mol. Biol. 75, 193–204. doi: 10.1007/s11103-010-9718-0
Yin, Y., Wang, Q., Xiao, L., Wang, F., Song, Z., Zhou, C., et al. (2018). Advances in the engineering of the gene editing enzymes and the genomes: understanding and handling the off-target effects of CRISPR/Cas9. J. Biomed. Nanotechnol. 14, 456–476. doi: 10.1166/jbn.2018.2537
Young, C. A., Tapper, B. A., May, K., Moon, C. D., Schardl, C. L., and Scott, B. (2009). Indole-diterpene biosynthetic capability of Epichloë endophytes as predicted by ltm gene analysis. J. Appl. Environ. Microbiol. 75, 2200–2211. doi: 10.1128/AEM.00953-08
Zhou, X., Jacobs, T. B., Xue, L. J., Harding, S. A., and Tsai, C. J. (2015). Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol. 208, 298–301. doi: 10.1111/nph.13470
Keywords: gene editing, New Zealand, regulation, traits, industry
Citation: Fritsche S, Poovaiah C, MacRae E and Thorlby G (2018) A New Zealand Perspective on the Application and Regulation of Gene Editing. Front. Plant Sci. 9:1323. doi: 10.3389/fpls.2018.01323
Received: 13 July 2018; Accepted: 22 August 2018;
Published: 12 September 2018.
Edited by:
Joachim Hermann Schiemann, Julius Kühn-Institut, GermanyReviewed by:
Drew Lloyd Kershen, University of Oklahoma, United StatesKan Wang, Iowa State University, United States
Copyright © 2018 Fritsche, Poovaiah, MacRae and Thorlby. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Glenn Thorlby, glenn.thorlby@scionresearch.com
 Steffi Fritsche
Steffi Fritsche Charleson Poovaiah
Charleson Poovaiah Elspeth MacRae
Elspeth MacRae Glenn Thorlby
Glenn Thorlby