Exposure to Diflubenzuron Results in an Up-Regulation of a Chitin Synthase 1 Gene in Citrus Red Mite, Panonychus citri (Acari: Tetranychidae)
Abstract
:1. Introduction
2. Results
2.1. Analyses of the cDNA and Protein Sequence of PcCHS1
2.2. Phylogenetic Analysis of PcCHS1
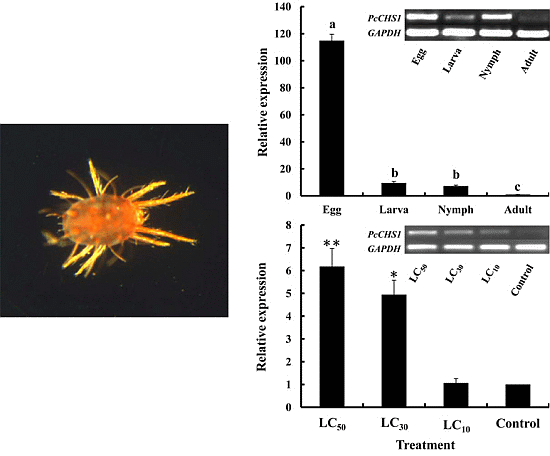
2.3. Development-Specific Expression Patterns of PcCHS1
2.4. Effects of DFB Treatment on the Expression of PcCHS1
3. Discussion
4. Experimental Section
4.1. P. Citri, Leaf-dip Bioassay
4.2. RNA Isolation and Reverse Transcription
4.3. Cloning and Sequencing of PcCHS1
4.4. Sequence Retrieval and Phylogenetic Analysis
4.5. Expression Patterns of PcCHS1 in Various P. citri Developmental Stages
4.6. Expression of PcCHS1 after DFB Exposure
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lehane, M. Peritrophic matrix structure and function. Annu. Rev. Entomol. 1997, 42, 525–550. [Google Scholar]
- Arakane, Y.; Hogenkamp, D.G.; Zhu, Y.C.; Kramer, K.J.; Specht, C.A.; Beeman, R.W.; Kanost, M.R.; Muthukrishnan, S. Characterization of two chitin synthase genes of the red flour beetle Tribolium castaneum and alternate exon usage in one of the genes during development. Insect Biochem. Mol. Biol. 2004, 34, 291–304. [Google Scholar]
- Kramer, K.; Muthukrishnan, S. Chitin metabolism in insects. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatrou, K., Gill, S., Eds.; Elsevier Press: Oxford, UK, 2005; Volume 4, pp. 111–144. [Google Scholar]
- Nagahashi, S.; Sudoh, M.; Ono, N.; Sawada, R.; Yamaguchi, E.; Uchida, Y.; Mio, T.; Takagi, M.; Arisawa, M.; Yamada-Okabe, H. Characterization of chitin synthase 2 of Saccharomyces cerevisiae Implication of two highly conserved domains as possible catalytic sites. J. Biol. Chem. 1995, 270, 13961–13967. [Google Scholar]
- Merzendorfer, H. Insect chitin synthases: A review. J. Comp. Physiol. B 2006, 176, 1–15. [Google Scholar]
- Tellam, R.L.; Vuocolo, T.; Johnson, S.E.; Jarmey, J.; Pearson, R.D. Insect chitin synthase—cDNA sequence gene organization and expression. Eur. J. Biochem 2000, 267, 6025–6042. [Google Scholar]
- Ibrahim, G.H.; Smartt, C.T.; Kiley, L.M.; Christensen, B.M. Cloning and characterization of a chitin synthase cDNA from the mosquito Aedes aegypti. Insect Biochem. Mol. Biol 2000, 30, 1213–1222. [Google Scholar]
- Zhang, X.; Zhang, J.Z.; Park, Y.; Zhu, K.Y. Identification and characterization of two chitin synthase genes in African malaria mosquito Anopheles gambiae. Insect Biochem. Mol. Biol. 2012, 42, 674–682. [Google Scholar]
- Zhang, J.Z.; Zhu, K.Y. Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron. Insect Biochem. Mol. Biol. 2006, 36, 712–725. [Google Scholar]
- Yang, W.J.; Xu, K.K.; Cong, L.; Wang, J.J. Identification mRNA expression and functional analysis of chitin synthase 1 gene and its two alternative splicing variants in oriental fruit fly Bactrocera dorsalis. Int. J. Biol. Sci. 2013, 9, 331. [Google Scholar]
- Ampasala, D.R.; Zheng, S.; Zhang, D.; Ladd, T.; Doucet, D.; Krell, P.J.; Retnakaran, A.; Feng, Q. An epidermis-specific chitin synthase CDNA in Choristoneura fumiferana: cloning characterization developmental and hormonal-regulated expression. Arch. Insect Biochem. Physiol. 2011, 76, 83–96. [Google Scholar]
- Gagou, M.E.; Kapsetaki, M.; Turberg, A.; Kafetzopoulos, D. Stage-specific expression of the chitin synthase DmeChSA and DmeChSB genes during the onset of Drosophila metamorphosis. Insect Biochem. Mol. Biol. 2002, 32, 141–146. [Google Scholar]
- Liang, Y.R.; Lin, C.; Wang, R.R.; Ye, J.H.; Lu, J.-L. Cloning and expression pattern of chitin synthase (CHS) gene in epidermis of Ectropis obliqua. Prout African J. Biotech. 2010, 9, 5297–5308. [Google Scholar]
- Zhang, J.Z.; Liu, X.J.; Zhang, J.P.; Li, D.Q.; Sun, Y.; Guo, Y.P.; Ma, E.; Zhu, K.Y. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust Locusta migratoria manilensis (Meyen). Insect Biochem. Mol. Biol. 2010, 40, 824–833. [Google Scholar]
- Hogenkamp, D.G.; Arakane, Y.; Zimoch, L.; Merzendorfer, H.; Kramer, K.J.; Beeman, R.W.; Kanost, M.R.; Specht, C.A.; Muthukrishnan, S. Chitin synthase genes in Manduca sexta: Characterization of a gut-specific transcript and differential tissue expression of alternately spliced mRNAs during development. Insect Biochem. Mol. Biol. 2005, 35, 529–540. [Google Scholar]
- Qu, M.B.; Yang, Q. A novel alternative splicing site of class A chitin synthase from the insect Ostrinia furnacalis—Gene organization expression pattern and physiological significance. Insect Biochem. Mol. Biol. 2011, 41, 923–931. [Google Scholar]
- Ashfaq, M.; Sonoda, S.; Tsumuki, H. Developmental and tissue-specific expression of CHS1 from Plutella xylostella and its response to chlorfluazuron. Pestic. Biochem. Physiol. 2007, 89, 20–30. [Google Scholar]
- Chen, X.; Yang, X.; Senthil Kumar, N.; Tang, B.; Sun, X.; Qiu, X.; Hu, J.; Zhang, W. The class A chitin synthase gene of Spodoptera exigua: Molecular cloning and expression patterns. Insect Biochem. Mol. Biol. 2007, 37, 409–417. [Google Scholar]
- Bolognesi, R.; Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Terra, W.R.; Ferreira, C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2005, 35, 1249–1259. [Google Scholar]
- Van Leeuwen, T.; Demaeght, P.; Osborne, E.J.; Dermauw, W.; Gohlke, S.; Nauen, R.; Grbic, M.; Tirry, L.; Merzendorfer, H.; Clark, R.M. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. USA 2012, 109, 4407–4412. [Google Scholar]
- Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Specht, C.A.; Tomoyasu, Y.; Lorenzen, M.D.; Kanost, M.; Beeman, R.W. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Biochem. Mol. Biol. 2005, 14, 453–463. [Google Scholar]
- Zimoch, L.; Merzendorfer, H. Immunolocalization of chitin synthase in the tobacco hornworm. Cell Tissue Res. 2002, 308, 287–297. [Google Scholar]
- Wang, Y.; Fan, H.W.; Huang, H.J.; Xue, J.; Wu, W.J.; Bao, Y.Y.; Xu, H.J.; Zhu, Z.R.; Cheng, J.A.; Zhang, C.X. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 2012, 42, 637–646. [Google Scholar]
- Bansal, R.; Mian, M.R.; Mittapalli, O.; Michel, A.P. Characterization of a chitin synthase encoding gene and effect of diflubenzuron in soybean aphid Aphis glycines. Int. J. Biol. Sci. 2012, 8, 1323. [Google Scholar]
- Silva, C.P.; Silva, J.R.; Vasconcelos, F.F.; Petretski, M.l.D.; DaMatta, R.A.; Ribeiro, A.F.; Terra, W.R. Occurrence of midgut perimicrovillar membranes in paraneopteran insect orders with comments on their function and evolutionary significance. Arthropod Struct. Dev. 2004, 33, 139–148. [Google Scholar]
- Gotoh, T.; Ishikawa, Y.; Kitashima, Y. Life-history traits of the six Panonychus species from Japan (Acari: Tetranychidae). Exp. Appl. Acarol 2003, 29, 241–252. [Google Scholar]
- Vassiliou, V.A.; Papadoulis, G. First record of the citrus red mite Panonychus citri in Cyprus. Phytoparasitica 2009, 37, 99–100. [Google Scholar]
- Ding, T.B.; Niu, J.Z.; Yang, L.H.; Zhang, K.; Dou, W.; Wang, J.J. Transcription profiling of two cytochrome P450 genes potentially involved in acaricide metabolism in citrus red mite Panonychus citri. Pestic. Biochem. Physiol. 2013, 106, 28–37. [Google Scholar]
- Zhang, K.; Niu, J.Z.; Ding, T.B.; Dou, W.; Wang, J.J. Molecular characterization of two Carboxylesterase genes of the citrus red mite Panonychus citri (Acari: Tetranychidae). Arch. Insect Biochem. Physiol. 2013, 82, 213–226. [Google Scholar]
- Whalon, M.; Mota-Sanchez, D.; Hollingworth, R.; Duynslager, L. Arthropod Pesticide Resistance Database(APRD). Available online: http://www.pesticideresistance.org (accessed on 1 August 2013).
- Meola, S.M.; Mayer, R.T. Inhibition of cellular proliferation of imaginal epidermal cells by diflubenzuron in pupae of the Stable fly. Science 1980, 207, 985–987. [Google Scholar]
- Mayer, R.; Chen, A.; DeLoach, J. Chitin synthesis inhibiting insect growth regulators do not inhibit chitin synthase. Experientia 1981, 37, 337–338. [Google Scholar]
- Merzendorfer, H.; Kim, H.S.; Chaudhari, S.S.; Kumari, M.; Specht, C.A.; Butcher, S.; Brown, S.J.; Robert Manak, J.; Beeman, R.W.; Kramer, K.J. Genomic and proteomic studies on the effects of the insect growth regulator diflubenzuron in the model beetle species Tribolium castaneum. Insect Biochem. Mol. Biol. 2012, 42, 264–276. [Google Scholar]
- Deul, D.; de Jong, B.; Kortenbach, J. Inhibition of chitin synthesis by two 1-(26-disubstituted benzoyl)-3-phenylurea insecticides II. Pestic. Biochem. Physiol. 1978, 8, 98–105. [Google Scholar]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar]
- Zimoch, L.; Hogenkamp, D.; Kramer, K.; Muthukrishnan, S.; Merzendorfer, H. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochem. Mol. Biol. 2005, 35, 515–527. [Google Scholar]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar]
- Rezende, G.L.; Martins, A.J.; Gentile, C.; Farnesi, L.C.; Pelajo-Machado, M.; Peixoto, A.A.; Valle, D. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle. BMC Dev. Biol. 2008, 8, 82. [Google Scholar]
- Zhang, X.; Zhang, J.; Zhu, K. Chitosan/double—Stranded RNA nanoparticle mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar]
- Qu, M.B.; Yang, Q. Physiological significance of alternatively spliced exon combinations of the single-copy gene class A chitin synthase in the insect Ostrinia furnacalis (Lepidoptera). Insect Mol. Biol. 2012, 21, 395–404. [Google Scholar]
- Grbić, M.; van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouzé, P.; Grbić, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.T.; Ortego, F. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar]
- Mulder, R.; Gijswijt, M.J. The laboratory evaluation of two promising new insecticides which interfere with cuticle deposition. Pest Manag. Sci. 1973, 4, 737–745. [Google Scholar]
- Clarke, B.S.; Jewess, P.J. The inhibition of chitin synthesis in Spodoptera littoralis larvae by flufenoxuron teflubenzuron and diflubenzuron. Pest Manag. Sci. 1990, 28, 377–388. [Google Scholar]
- Hajjar, N.P.; Casida, J.E. Insecticidal benzoylphenyl ureas: Structure-activity relationships as chitin synthesis inhibitors. Science 1978, 200, 1499–1500. [Google Scholar]
- Gangishetti, U.; Breitenbach, S.; Zander, M.; Saheb, S.K.; Müller, U.; Schwarz, H.; Moussian, B. Effects of benzoylphenylurea on chitin synthesis and orientation in the cuticle of the Drosophila larva. Eur. J. Cell. Biol 2009, 88, 167–180. [Google Scholar]
- Takafuji, Y.; Fujimoto, H. Winter survival of the non-diapausing population of the citrus red mite Panonychus citri (MaGregor) (Acarian: Tetranychidae) on pear and citrus. Appl. Entomol. Zool. 1986, 21, 467–473. [Google Scholar]
- Arakane, Y.; Specht, C.A.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Chitin synthases are required for survival fecundity and egg hatch in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 959–962. [Google Scholar]
- Khila, A.; Grbić, M. Gene silencing in the spider mite Tetranychus urticae: dsRNA and siRNA parental silencing of the Distal-less gene. Dev. Genes. Evol. 2007, 217, 241–251. [Google Scholar]
- Kwon, D.H.; Park, J.H.; Lee, S.H. Screening of lethal genes for feeding RNAi by leaf disc-mediated systematic delivery of dsRNA in Tetranychus urticae. Pestic. Biochem. Physiol. 2012, 105, 69–75. [Google Scholar]
- Hu, J.; Wang, C.; Wang, J.; You, Y.; Chen, F. Monitoring of resistance to spirodiclofen and five other acaricides in Panonychus citri collected from Chinese citrus orchards. Pest Manag. Sci. 2010, 66, 1025–1030. [Google Scholar]
- Michel, A.P.; Mian, M.R.; Davila-Olivas, N.H.; Cañas, L.A. Detached leaf and whole plant assays for soybean aphid resistance: differential responses among resistance sources and biotypes. J. Econ. Entomol. 2010, 103, 949–957. [Google Scholar]
- Niu, J.Z.; Dou, W.; Ding, T.B.; Shen, G.M.; Zhang, K.; Smagghe, G.; Wang, J.J. Transcriptome analysis of the citrus red mite Panonychus citri and its gene expression by exposure to insecticide/acaricide. Insect Mol. Biol. 2012, 21, 422–436. [Google Scholar]
- Hill, C.B.; Hartman, G.L. Resistance to the soybean aphid in soybean germplasm. Crop Sci 2004, 44, 98–106. [Google Scholar]
- Bansal, R.; Hulbert, S.; Schemerhorn, B.; Reese, J.C.; Whitworth, R.J.; Stuart, J.J.; Chen, M.S. Hessian fly-associated bacteria: transmission essentiality and composition. PLoS One 2011, 6, e23170. [Google Scholar]
- Bairoch, A. The PROSITE dictionary of sites and patterns in proteins its current status. Nucleic Acids. Res 1993, 21, 3097–3103. [Google Scholar]
- Bendtsen, J.D.; Nielsen, H.; von Heijing, G.; Brunak, S. Improved prediction os signal preptides: SignalIP 30. J. Mol. Biol 2004, 340, 783–795. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijine, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol 2001, 305, 567–580. [Google Scholar]
- Hansen, J.E.; Lund, O.; Rapachi, K.; Brunak, S. O-GLYCBASE versin 20: A revised database of O-glycosylated proteins. Nucleic. Acids. Res 1997, 25, 278–282. [Google Scholar]
- Tamura, K.; Peyerson, D.; Peterson, N.; Stecher, G.; Nei Kumar, S. MAGA5: Molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol. Biol. Evol 2011, 28, 2731–2739. [Google Scholar]
- Rozen, S.; Skaletsky, H. Primer 3 on the WWW for General user and for biologist programmers. Bioinforma. Methods Protoc. 1999, 132, 365–386. [Google Scholar]
- Niu, J.Z.; Dou, W.; Ding, T.B.; Yang, L.H.; Shen, G.M.; Wang, J.J. Evaluation of suitable reference genes for quantitative RT-PCR during development and abiotic stress in Panonychus citri (McGregor) (Acari: Tetranychidae). Mol. Biol. Rep. 2012, 39, 5841–5849. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]







| Genes | GenBank No. or Gene ID. | Species |
|---|---|---|
| CHS1 | XP001662200 | Aedes aegypti |
| CHS1 | XP_321336 | Anopheles gambiae |
| CHS1 | XP_395677 | Apis mellifera |
| CHS1 | XP_001866798 | Culex quinquefasciatus |
| CHS1 | NM_079509 | Drosophila melanogaster |
| CHS1 | XP_001359390 | Daphnia pulex |
| CHS1 | ACD10533 | Ectropis oblique Prout |
| CHS1 | AF221067 | Lucilia cuprina |
| CHS1 | GU067731 | Locusta migratoria manilensis |
| CHS1 | JQ040011 | Laodelphax striatella |
| CHS1 | AY062175 | Manduca sexta |
| CHS1 | BAF47974 | Plutella xylostella |
| CHS1 | DQ062153 | Spodoptera exigua |
| CHS1 | AY291476 | Tribolium castaneum |
| CHS1 | tetur03g08510 | Tetranychus urticae |
| CHS1 | KF241748 | Panonychus citri |
| CHS1 | XP003741992 | Metaseiulus occidentalis |
| CHS2 | XP_001651163 | Aedes aegypti |
| CHS2 | XP_321951 | Anopheles gambiae |
| CHS2 | XP_001121152 | Apis mellifera |
| CHS2 | XP_001864594 | Culex quinquefasciatus |
| CHS2 | NM_079485 | Drosophila melanogaster |
| CHS2 | EFX80669 | Daphnia pulex |
| CHS2 | AAX20091 | Manduca sexta |
| CHS2 | ABB97082 | Ostrinia furnacalis |
| CHS2 | XP_002423604 | Pediculus humanus corporis |
| CHS2 | EU622827 | Spodoptera exigua |
| CHS2 | AY525599 | Spodoptera frugiperda |
| CHS2 | AY291477 | Tribolium castaneum |
| CHS2 | tetur08g00170 | Tetranychus urticae |
| Experiments | Primer names and sequences (5′ to 3′) | Product length (bp) |
|---|---|---|
| PcCHS1-1-S: GATGTGACCAAGGGCGTAA | 1717 | |
| PcCHS1-1-A: CGCCGAGATTTTTATTTTTC | ||
| Specific PCR | PcCHS1-2-S: CGGAAGCCGTTCAACTATTA | 1324 |
| PcCHS1-2-A: TGTTCCTCGTCTTCCGTCAC | ||
| PcCHS1-3-S: GTTACCTGGGGAACTCGTGA | 1173 | |
| PcCHS1-3-A: TTGTTGAGCGGCTGAAAGTT | ||
| PcCHS1b-S: TATCTCGCCGATATTGAGGTC | 367 | |
| PcCHS1b-A: GATGGAAAAGCATACCGACCA | ||
| 3′ RACE | PcCHS1-S1: ATGCTCTTTTCGTTCTCGTG | 722 |
| PcCHS1-S2: TTGTCGGTTTCTTTGGATTA | 560 | |
| 5′ RACE | PcCHS1-A1: TTGGTCACATCCAGCGAAG | 503 |
| PcCHS1-A2: TATCCTGGCAGCACGAA | 375 | |
| Full-length confirmation | PcCHS1-1: TAACATCCAAATGGGTGCTGA | 1000 |
| PcCHS1-2: GAACCATCAACCAAAAGTCGG | ||
| PcCHS1-3: CGCTGAATCAATCGGTTTAGGT | 1100 | |
| PcCHS1-4: TATCGCTGAAGCAGATGAACGC | ||
| PcCHS1-5: ATTTACTTTCCATCGGTGCCCA | 3131 | |
| PcCHS1-6: CCACGAGAACGAAAAGAGCATT | ||
| PcCHS1 | PcCHS1-Q-F: AAGTTTGAATACGCGGTTGG | 191 |
| PcCHS1-Q-R: CGATCTTCCCCTTGATCGTA | ||
| GAPDH | GAPDH-F: CTTTGGCCAAGGTCATCAAT | 159 |
| GAPDH-R: CGGTAGCGGCAGGTATAATG | ||
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xia, W.-K.; Ding, T.-B.; Niu, J.-Z.; Liao, C.-Y.; Zhong, R.; Yang, W.-J.; Liu, B.; Dou, W.; Wang, J.-J. Exposure to Diflubenzuron Results in an Up-Regulation of a Chitin Synthase 1 Gene in Citrus Red Mite, Panonychus citri (Acari: Tetranychidae). Int. J. Mol. Sci. 2014, 15, 3711-3728. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms15033711
Xia W-K, Ding T-B, Niu J-Z, Liao C-Y, Zhong R, Yang W-J, Liu B, Dou W, Wang J-J. Exposure to Diflubenzuron Results in an Up-Regulation of a Chitin Synthase 1 Gene in Citrus Red Mite, Panonychus citri (Acari: Tetranychidae). International Journal of Molecular Sciences. 2014; 15(3):3711-3728. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms15033711
Chicago/Turabian StyleXia, Wen-Kai, Tian-Bo Ding, Jin-Zhi Niu, Chong-Yu Liao, Rui Zhong, Wen-Jia Yang, Bin Liu, Wei Dou, and Jin-Jun Wang. 2014. "Exposure to Diflubenzuron Results in an Up-Regulation of a Chitin Synthase 1 Gene in Citrus Red Mite, Panonychus citri (Acari: Tetranychidae)" International Journal of Molecular Sciences 15, no. 3: 3711-3728. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms15033711