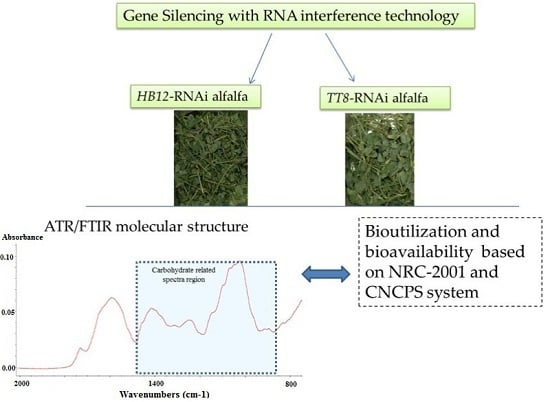
Gene-Silencing-Induced Changes in Carbohydrate Conformation in Relation to Bioenergy Value and Carbohydrate Subfractions in Modeled Plant (Medicago sativa) with Down-Regulation of HB12 and TT8 Transcription Factors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Changes in Nutrient Bioutlization and Bioavailability in Alfalfa with Down-Regulated TT8 and HB12 Genes
2.2. Effects of TT8 and HB12 Silencing on Carbohydrate Sub-Fractions in Alfalfa Forage
2.3. Multivariate Analysis of Carbohydrate Molecular Spectral Profiles that Are Affected by HB12 and TT8 Silencing
2.4. Relationship between Gene-Silencing-Induced Molecular Structure Changes and Nutrient Bioutilization and Bioavailability and Bioenergy of Alfalfa Forage
2.5. Relationship between Gene Silencing-Induced Molecular Structure Changes and Carbohydrate SubFractions in Alfalfa Forage Populations
2.6. Predictions for Gene-Silencing-Induced Molecular Structure Changes and Nutrient Availability of Alfalfa Forage
3. Experimental Section
3.1. HB12-RNAi, TT8-RNAi and Non-Transgenic Alfalfa Population Material
Generating RNAi Constructs and Transformation of Alfalfa
3.2. Advanced Non-Invasive Molecular Spectroscopy-FT/IR
3.3. Multivariate Molecular Spectral Analyses of CHO Spectral Data Collected from Alfalfa Forage (Medicago sativa) with Down-Regulation of TT8 and HB12 Genes
3.4. Evaluation of Nutrient BioUtilization and Bioavailability and BioEnergy Profiles in Alfalfa Forage (Medicago sativa) with Down-Regulation of TT8 and HB12 Genes
3.5. Partitioning Carbohydrate Subfractions in Alfalfa Forage (Medicago sativa) with Down-Regulation of TT8 and HB12 Genes
3.6. Statistical Analysis
3.6.1. Bioutilization and Bioavailability of Nutrients, Bioenergy Profiles and Carbohydrate Subfractions
3.6.2. Correlation Analysis of CHO Spectral Profiles with CHO Nutrient Supply
3.6.3. Multiple Regression Analysis of CHO Spectral Profiles with Carbohydrate Subfractions, Bioenergy Value and Digestible Nutrients
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jonker, A.; Gruber, M.Y.; McCaslin, M.; Wang, Y.; Coulman, B.; McKinnon, J.J.; Christensen, D.A.; Yu, P. Nutrient composition and degradation profiles of anthocyanidin-accumulating Lc-alfalfa populations. Can. J. Anim. Sci. 2010, 90, 401–412. [Google Scholar] [CrossRef]
- Jonker, A.; Gruber, M.Y.; Wang, Y.; Coulman, B.; Azarfar, A.; McKinnon, J.J.; Christensen, D.A.; Yu, P. Modeling degradation ratios and nutrient availability of anthocyanidin-accumulating Lc-Alfalfa populations in dairy cows. J. Dairy Sci. 2011, 94, 1430–1444. [Google Scholar] [CrossRef] [PubMed]
- Jonker, A.; Gruber, M.; Wang, Y.; Coulman, B.; McKinnon, J.J.; Christensen, D.; Yu, P. Foam stability of leaves from anthocyanidin-accumulating Lc-alfalfa and relation to molecular structures detected by fourier-transformed infrared-vibration spectroscopy. Grass Forage Sci. 2012, 67, 369–381. [Google Scholar] [CrossRef]
- Jonker, A.; Gruber, M.Y.; Wang, Y.; Narvaez, N.; Coulman, B.; McKinnon, J.J.; Christensen, D.A.; Azarfar, A.; Yu, P. Fermentation, degradation and microbial nitrogen partitioning for three forage color phenotypes within anthocyanidin-accumulating Lc-alfalfa progeny. J. Sci. Food Agric. 2012, 92, 2265–2273. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Jonker, A.; Gruber, M. Molecular basis of protein structure in proanthocyanidin and anthocyanin-enhanced Lc-transgenic alfalfa in relation to nutritive value using synchrotron-radiation FTIR microspectroscopy: A novel approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2009, 73, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Hannoufa, A.; Yu, P. Transformation with TT8 and HB12 RNAi constructs in model forage (Medicago sativa, alfalfa) affects carbohydrate structure and metabolic characteristics in dairy cattle. J. Agric. Food Chem. 2015, 63, 9590–9600. [Google Scholar] [CrossRef] [PubMed]
- Nookaraju, A.; Pandey, S.K.; Bae, H.J.; Joshi, C.P. Designing cell walls for improved bioenergy production. Mol. Plant 2013, 6, 8–10. [Google Scholar] [CrossRef] [PubMed]
- NRC. Nutrient Requirements of Dairy Cattle, 7th revised ed.; National Academy of Sciences: Washington, DC, USA, 2001; p. 381. [Google Scholar]
- NRC. Nutrient Requirements of Beef Cattle, 7th ed.; National Academy Press: Washington, DC, USA, 1996. [Google Scholar]
- Tylutki, T.; Fox, D.; Durbal, V.; Tedeschi, L.; Russell, J.; van Amburgh, M.; Overton, T.; Chase, L.; Pell, A. Cornell net carbohydrate and protein system: A model for precision feeding of dairy cattle. Anim. Feed Sci. Technol. 2008, 143, 174–202. [Google Scholar] [CrossRef]
- AMTS. Agricultural Modeling and Training Systems; Cornell Research Foundation: Ithaca, NY, USA, 2010. [Google Scholar]
- Zhang, X.; Yu, P. Relationship of carbohydrate molecular spectroscopic features in combined feeds to carbohydrate utilization and availability in ruminants. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 92, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Application of cluster analysis (CLA) in feed chemical imaging to accurately reveal structural-chemical features of feeds and plants within cellular dimension. J. Agric. Food Chem. 2005, 53, 2872–2880. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Applications of hierarchical cluster analysis (CLA) and principal component analysis (PCA) in feed structure and feed molecular chemistry research, using synchrotron-based Fourier transform infrared (FTIR) microspectroscopy. J. Agric. Food Chem. 2005, 53, 7115–7127. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Yu, P. Molecular clustering, interrelationships and carbohydrate conformation in hull and seeds among barley cultivars. J. Cereal Sci. 2011, 53, 379–383. [Google Scholar] [CrossRef]
- Yu, P.; Damiran, D.; Azarfar, A.; Niu, Z. Detecting molecular features of spectra mainly associated with structural and non-structural carbohydrates in co-products from bioethanol production using DRIFT with uni-and multivariate molecular spectral analyses. Int. J. Mol. Sci. 2011, 12, 1921–1935. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Short communication: Relationship of carbohydrate molecular spectroscopic features to carbohydrate nutrient profiles in co-products from bioethanol production. J. Dairy Sci. 2012, 95, 2091–2096. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Falk, K.C.; Yu, P. Studies on Brassica carinata seed. 2. Carbohydrate molecular structure in relation to carbohydrate chemical profile, energy values, and biodegradation characteristics. J. Agric. Food Chem. 2013, 61, 10127–10134. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, X.; Yu, P. Correlating molecular spectroscopy and molecular chemometrics to explore carbohydrate functional groups and utilization of coproducts from biofuel and biobrewing processing. J. Agric. Food Chem. 2014, 62, 5108–5117. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; McKinnon, J.J.; Christensen, D.A.; Beattie, A.D.; Yu, P. Characterizing the molecular structure features of newly developed hulless barley cultivars with altered carbohydrate traits (Hordeum vulgare L.) by globar-sourced infrared spectroscopy in relation to nutrient utilization and availability. J. Cereal Sci. 2014, 60, 48–59. [Google Scholar] [CrossRef]
- Badhan, A.; Jin, L.; Wang, Y.; Han, S.; Kowalczys, K.; Brown, D.C.W.; Ayala, C.J.; Latoszek-Green, M.; Miki, B.; Tsang, A.; et al. Expression of a fungal ferulic acid esterase in alfalfa modifies cell wall digestibility. Biotechnol. Biofuels 2014, 12, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.M.; Dumas, P. Chemical imaging of biological tissue with synchrotron infrared light. Biochim. Biophys. Acta 2006, 1758, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Christensen, C.R.; Christensen, D.A.; McKinnon, J.J. Ultrastructural-chemical makeup of yellow-seeded (Brassica rapa) and brown-seeded (Brassica napus) canola within cellular dimensions, explored with synchrotron reflection FTIR microspectroscopy. Can. J. Plant Sci. 2005, 85, 533–541. [Google Scholar] [CrossRef]
- Weiss, W.; Conrad, H.; Pierre, N.S. A theoretically-based model for predicting total digestible nutrient values of forages and concentrates. Anim. Feed Sci. Technol. 1992, 39, 95–110. [Google Scholar] [CrossRef]
- Lanzas, C.; Sniffen, C.; Seo, S.; Tedeschi, L.; Fox, D. A revised CNCPS feed carbohydrate fractionation scheme for formulating rations for ruminants. Anim. Feed Sci. Technol. 2007, 136, 167–190. [Google Scholar] [CrossRef]
- SAS. SAS User’s Guide, Version 9.3; SAS Institute Inc.: Cary, NC, USA, 2011. [Google Scholar]




| Items | Non-Transgenic (NT) | Gene Silencing through RNAi Technology (GS) | SEM b | p Value | Contrast, p Value | |
|---|---|---|---|---|---|---|
| Control | HB12 | TT8 | NT vs. GS | |||
| Truly digestible nutrient c (% DM) | ||||||
| tdNFC | 42.31 a | 38.68 b | 37.37 b | 0.539 | 0.016 | 0.007 |
| tdCPc | 20.82 b | 19.42 c | 22.34 a | 0.256 | 0.009 | 0.861 |
| tdFA | 0.57 | 0.88 | 0.45 | 0.131 | 0.197 | 0.605 |
| tdNDF | 14.49 | 14.78 | 14.89 | 0.622 | 0.898 | 0.679 |
| Total digestible nutrient d (% DM) | ||||||
| TDN1x | 71.89 | 67.85 | 68.59 | 1.099 | 0.149 | 0.072 |
| Predicted energy value e (Mcal/kg DM) | ||||||
| DE1x | 3.31 | 3.12 | 3.19 | 0.048 | 0.141 | 0.077 |
| NEm | 1.79 | 1.66 | 1.71 | 0.035 | 0.164 | 0.092 |
| NEg | 1.17 | 1.05 | 1.09 | 0.030 | 0.142 | 0.078 |
| DE3x | 3.04 | 2.86 | 2.93 | 0.044 | 0.143 | 0.081 |
| ME3x | 2.62 | 2.44 | 2.51 | 0.047 | 0.160 | 0.089 |
| NELP | 1.65 | 1.52 | 1.57 | 0.031 | 0.140 | 0.075 |
| Items | Non-Transgenic (NT) | Gene silencing through RNAi Technology (GS) | SEM b | p Value | Contrast, p Value | |
|---|---|---|---|---|---|---|
| Control | HB12 | TT8 | NT vs. GS | |||
| Fractions of Carbohydrate Partitioned by CNCPS c (% CHO) | ||||||
| CA4 | 7.37 | 4.18 | 4.41 | 0.867 | 0.135 | 0.063 |
| CB1 | 5.26 a | 2.04 b | 2.58 b | 0.334 | 0.012 | 0.006 |
| CB2 | 49.35 | 49.79 | 49.41 | 1.201 | 0.962 | 0.876 |
| CB3 | 29.89 | 29.64 | 31.30 | 1.534 | 0.735 | 0.779 |
| CC | 8.14 | 14.36 | 12.31 | 1.908 | 0.209 | 0.113 |
| Item a | TDN1x % DM | tdNFC % DM | tdFA % DM | tdNDF % DM | ||||
|---|---|---|---|---|---|---|---|---|
| r b | p Value | r | p Value | r | p Value | r | p Value | |
| Structural CHO related Spectral Profiles | ||||||||
| A_StCHO | −0.09 | 0.872 | 0.09 | 0.872 | −0.54 | 0.266 | −0.81 | 0.050 |
| H_1415 | 0.19 | 0.725 | 0.37 | 0.470 | −0.49 | 0.320 | −0.70 | 0.118 |
| H_1370 | −0.06 | 0.913 | 0.12 | 0.827 | −0.61 | 0.200 | −0.87 | 0.025 |
| H_1315 | 0.00 | 1.000 | 0.09 | 0.868 | −0.71 | 0.117 | −0.85 | 0.032 |
| A_1315 | −0.83 | 0.042 | −0.83 | 0.042 | −0.14 | 0.787 | −0.32 | 0.538 |
| Cellulosic Compounds Related Spectral Profiles | ||||||||
| H_1244 | 0.09 | 0.868 | 0.00 | 1.000 | −0.85 | 0.031 | −0.72 | 0.109 |
| A_CELC | 0.09 | 0.872 | −0.09 | 0.872 | −0.83 | 0.042 | −0.55 | 0.257 |
| Total CHO related Spectral Profiles | ||||||||
| A_CHO | 0.03 | 0.957 | 0.03 | 0.957 | −0.71 | 0.111 | −0.75 | 0.084 |
| H_1150 | −0.03 | 0.957 | 0.06 | 0.913 | −0.64 | 0.173 | −0.79 | 0.059 |
| H_1100 | −0.03 | 0.957 | 0.06 | 0.913 | −0.64 | 0.173 | −0.79 | 0.059 |
| H_1025 | −0.03 | 0.957 | 0.06 | 0.913 | −0.64 | 0.173 | −0.79 | 0.059 |
| Non-structural CHO related Spectral Profiles | ||||||||
| A_non-st CHO | −0.09 | 0.872 | 0.09 | 0.872 | −0.54 | 0.266 | −0.81 | 0.500 |
| H_895 | −0.13 | 0.805 | 0.39 | 0.441 | 0.13 | 0.805 | −0.66 | 0.150 |
| Spectral Ratio Profiles | ||||||||
| A_StCHO/A_CHO ratio | 0.49 | 0.329 | 0.71 | 0.111 | −0.09 | 0.872 | −0.26 | 0.618 |
| A_Non-stCHO/A_CHO ratio | −0.41 | 0.414 | 0.00 | 1.000 | 0.83 | 0.042 | 0.11 | 0.843 |
| A_Non-stCHO/A_StCHO ratio | −0.21 | 0.695 | 0.03 | 0.956 | 0.91 | 0.011 | 0.40 | 0.428 |
| A_CELC/A_CHO ratio | −0.03 | 0.957 | −0.43 | 0.397 | −0.94 | 0.005 | −0.32 | 0.538 |
| A_CELC/A_StCHO ratio | −0.09 | 0.872 | −0.54 | 0.266 | −0.54 | 0.266 | 0.06 | 0.913 |
| Item a | CA4 % CHO | CB1 % CHO | CB2 % CHO | CB3 % CHO | CC % CHO | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| r b | p Value | r | p Value | r | p Value | r | p Value | r | p Value | |
| Structural CHO related Spectral Profiles | ||||||||||
| A_StCHO | 0.71 | 0.111 | 0.71 | 0.111 | −0.60 | 0.208 | −0.77 | 0.072 | 0.09 | 0.872 |
| H_1415 | 0.83 | 0.039 | 0.83 | 0.039 | −0.62 | 0.192 | −0.62 | 0.192 | −0.19 | 0.725 |
| H_1370 | 0.75 | 0.084 | 0.75 | 0.084 | −0.70 | 0.125 | −0.81 | 0.050 | 0.06 | 0.913 |
| H_1315 | 0.79 | 0.059 | 0.79 | 0.059 | −0.62 | 0.191 | −0.79 | 0.059 | 0.00 | 1.000 |
| A_1315 | −0.43 | 0.397 | −0.43 | 0.397 | −0.14 | 0.787 | −0.43 | 0.397 | 0.83 | 0.042 |
| Cellulosic Compounds Related Spectral Profiles | ||||||||||
| H_1244 | 0.79 | 0.059 | 0.79 | 0.059 | −0.47 | 0.346 | −0.65 | 0.165 | −0.09 | 0.868 |
| A_CELC | 0.71 | 0.111 | 0.71 | 0.111 | −0.31 | 0.544 | −0.49 | 0.329 | −0.09 | 0.872 |
| Total CHO related Spectral Profiles | ||||||||||
| A_CHO | 0.77 | 0.072 | 0.77 | 0.072 | −0.43 | 0.397 | −0.71 | 0.111 | -0.03 | 0.957 |
| H_1150 | 0.75 | 0.084 | 0.75 | 0.084 | −0.52 | 0.288 | −0.75 | 0.084 | 0.03 | 0.957 |
| H_1100 | 0.75 | 0.084 | 0.75 | 0.084 | −0.52 | 0.288 | −0.75 | 0.084 | 0.03 | 0.957 |
| H_1025 | 0.75 | 0.084 | 0.75 | 0.084 | −0.52 | 0.288 | −0.75 | 0.084 | 0.03 | 0.957 |
| Non-structural CHO related Spectral Profiles | ||||||||||
| A_non-st CHO | 0.71 | 0.111 | 0.71 | 0.111 | -0.60 | 0.208 | −0.77 | 0.072 | 0.09 | 0.872 |
| H_895 | 0.39 | 0.441 | 0.39 | 0.441 | -0.65 | 0.158 | −0.65 | 0.158 | 0.13 | 0.805 |
| Spectral Ratio Profiles | ||||||||||
| A_StCHO/A_CHO ratio | 0.66 | 0.156 | 0.66 | 0.156 | −0.43 | 0.397 | −0.14 | 0.787 | −0.49 | 0.329 |
| A_Non-stCHO/A_CHO ratio | −0.62 | 0.188 | −0.62 | 0.188 | 0.00 | 1.000 | 0.00 | 1.000 | 0.41 | 0.414 |
| A_Non-stCHO/A_StCHO ratio | −0.65 | 0.165 | −0.65 | 0.165 | 0.50 | 0.312 | 0.26 | 0.612 | 0.21 | 0.695 |
| A_CELC/A_CHO ratio | 0.37 | 0.469 | 0.37 | 0.469 | −0.20 | 0.704 | −0.26 | 0.623 | 0.03 | 0.957 |
| A_CELC/A_StCHO ratio | 0.03 | 0.957 | 0.03 | 0.957 | 0.37 | 0.469 | 0.03 | 0.957 | 0.09 | 0.872 |
| Predicted Variables (Y) | Variable Selection (Variables Left in the Model with p < 0.05) | Prediction Equations (Test Model: Y = a + b1x1 + b2x2) | Model R2 Value | RSD a | p Value |
|---|---|---|---|---|---|
| Carbohydrate fraction profiles | |||||
| CA4 (% CHO) | Ratio of structural CHO to total CHO left in the model | Y = −41.04 + 99.32 A_StCHO/A_CHO ratio | 0.77 | 1.00 | 0.022 |
| CB1 (% CHO) | H_1415 left in the model | Y = −6.91 + 355.77 H_1415 | 0.70 | 0.97 | 0.038 |
| CB3 (% CHO) | H_895 left in the model | Y = 42.57 − 4338.00 H_895 | 0.91 | 0.64 | 0.004 |
| Total digestible nutrients | |||||
| tdNDF (% DM) | A_non-STCHO left in the model | Y = 21.84 − 83.78 A_non-STCHO | 0.88 | 0.28 | 0.006 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Hannoufa, A.; Zhang, Y.; Yu, P. Gene-Silencing-Induced Changes in Carbohydrate Conformation in Relation to Bioenergy Value and Carbohydrate Subfractions in Modeled Plant (Medicago sativa) with Down-Regulation of HB12 and TT8 Transcription Factors. Int. J. Mol. Sci. 2016, 17, 720. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050720
Li X, Hannoufa A, Zhang Y, Yu P. Gene-Silencing-Induced Changes in Carbohydrate Conformation in Relation to Bioenergy Value and Carbohydrate Subfractions in Modeled Plant (Medicago sativa) with Down-Regulation of HB12 and TT8 Transcription Factors. International Journal of Molecular Sciences. 2016; 17(5):720. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050720
Chicago/Turabian StyleLi, Xinxin, Abdelali Hannoufa, Yonggen Zhang, and Peiqiang Yu. 2016. "Gene-Silencing-Induced Changes in Carbohydrate Conformation in Relation to Bioenergy Value and Carbohydrate Subfractions in Modeled Plant (Medicago sativa) with Down-Regulation of HB12 and TT8 Transcription Factors" International Journal of Molecular Sciences 17, no. 5: 720. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050720