Phytocystatins: Defense Proteins against Phytophagous Insects and Acari
Abstract
:1. Phytocystatin Features
2. Phytocystatin Functions
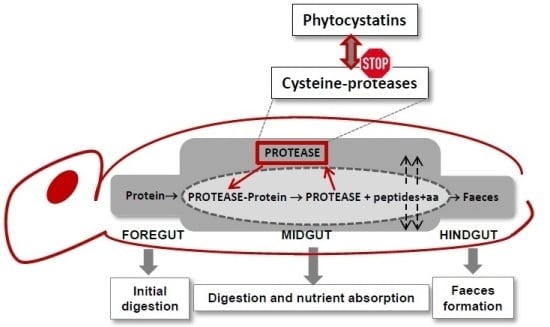
3. Phytocystatin Targets: Arthropod Proteases
4. Tailoring and Selection of Phytocystatin for Pest Control
5. Arthropod Herbivore Responses and Adaptations
6. Biotechnological Cystatin Applications to Control Pests
6.1. Recombinant Phytocystatins for in Vitro and in Vivo Assays
6.2. Phytocystatins and Transgenic Plants
7. Conclusions and Future
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, M.E.; Diaz-Mendoza, M.; Diaz, I.; Martinez, M. Plant protein peptidase inhibitors: An evolutionary overview based on comparative genomics. BMC Genom. 2014, 15, 812. [Google Scholar] [CrossRef] [PubMed]
- Kordis, D.; Turk, V. Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol. Biol. 2009, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Benchabane, M.; Schlüter, U.; Vorster, J.; Goulet, M.C.; Michaud, D. Plant cystatins. Biochimie 2010, 92, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Diaz-Mendoza, M.; Carrillo, L.; Diaz, I. Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and legumain cysteine proteinases. FEBS Lett. 2007, 581, 2914–2918. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Diaz, I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol. Biol. 2008, 8, 198. [Google Scholar] [CrossRef] [PubMed]
- Green, A.R.; Nissen, M.S.; Kumar, G.N.M.; Knowles, N.R.; Kang, C.H. Characterization of Solanum tuberosum multicystatin and the significance of core domains. Plant Cell 2013, 25, 5043–5052. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Kudo, N.; Abe, K.; Arai, S.; Tanokura, M. Three-dimensional solution structure of oryzacystatin-I; a cysteine proteinase inhibitor of the rice; Oryza sativa L. japonica. Biochemistry 2000, 39, 14753–14760. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.H.; Liu, K.L.; Wu, H.Y.; Yeh, K.W.; Cheng, Y.S. Crystal structure of tarocystatin-papain complex: Implications for the inhibition property of group-2 phytocystatins. Planta 2011, 23, 4243–4254. [Google Scholar] [CrossRef] [PubMed]
- Irene, D.; Chung, T.Y.; Chen, B.J.; Liu, T.H.; Li, F.Y.; Tzen, J.T.; Wang, C.I.; Chyan, C.L. Solution structure of a phytocystatin from Ananas comosus and its molecular interaction with papain. PLoS ONE 2012, 7, e47865. [Google Scholar] [CrossRef] [PubMed]
- Valadares, N.F.; de Oliveira-Silva, R.; Cavini, I.A.; Marques Ide, A.; Pereira, H.D.; Soares-Costa, A.; Henrique-Silva, F.; Kalbitzer, H.R.; Munte, C.E.; Garratt, R.C. X-ray crystallography and NMR studies of domain-swapped canecystatin-1. FEBS J. 2013, 280, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Rasoolizadeh, A.; Goulet, M.C.; Sainsbury, F.; Cloutier, C.; Michaud, D. Single substitutions to closely related amino acids contribute to the functional diversification of an insect-inducible, positively selected plant cystatin. FEBS J. 2016, 283, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Van Wyk, S.G.; Kunert, K.J.; Cullis, C.A.; Pillay, P.; Makgopa, M.E.; Schlüter, U.; Vorster, B.J. Review: The future of cystatin engineering. Plant Sci. 2016, 246, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Cambra, I.; Carrillo, L.; Diaz-Mendoza, M.; Diaz, I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 2009, 151, 1531–1545. [Google Scholar] [CrossRef] [PubMed]
- Munger, A.; Simon, M.A.; Khalf, M.; Goulet, M.C.; Michaud, D. Cereal cystatins delay sprouting and nutrient loss in tubers of potato; Solanum tuberosum. BMC Plant Biol. 2015, 15, 296. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Mendoza, M.; Dominguez-Figueroa, J.D.; Velasco-Arroyo, B.; Cambra, I.; Gonzalez-Melendi, P.; Lopez-Gonzalvez, A.; Garcia, A.; Hensel, G.; Kumlehn, J.; Diaz, I.; et al. HvPap-1 C1A protease and HvCPI-2 cystatin contribute to barley grain filling and germination. Plant Physiol. 2016, 170, 2511–2524. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Mendoza, M.; Arroyo-Velasco, B.; Gonzalez-Melendi, P.; Martinez, M.; Diaz, I. C1A Cysteine protease-cystatin interactions in leaf senescence. J. Exp. Bot. 2014, 65, 3825–3833. [Google Scholar] [CrossRef] [PubMed]
- Kunert, K.J.; van Wyk, S.G.; Cullis, C.A.; Vorster, B.J.; Foyer, C.H. Potential use of phytocystatins in crop improvement; with a particular focus on legumes. J. Exp. Bot. 2015, 66, 3559–3570. [Google Scholar] [CrossRef] [PubMed]
- Bolter, C.J. Methyl jasmonate induces papain inhibitor(s) in tomato leaves. Plant Physiol. 1993, 103, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Botella, M.A.; Xu, Y.; Prabha, T.N.; Zhao, Y.; Narasimhan, M.L.; Wilson, K.A.; Nielsen, S.S.; Bressan, R.A.; Hasegawa, P.M. Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol. 1996, 112, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Pernas, M.; Sánchez-Monge, R.; Salcedo, G. Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett. 2000, 467, 206–210. [Google Scholar] [CrossRef]
- Van der Linde, K.; Hemetsberger, C.; Kastner, C.; Kaschani, F.; van der Hoorn, R.A.L.; Kumlehn, J.; Doehlemanna, G. A Maize Cystatin Suppresses Host Immunity by Inhibiting Apoplastic Cysteine Proteases. Plant Cell 2012, 24, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Dutt, S.; Gaur, V.S.; Taj, G.; Kumar, A. Differential induction of two different cystatin genes during pathogenesis of Karnal bunt (Tilletia indica) in wheat under the influence of jasmonic acid. Gene 2012, 506, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; D’Alessandro, M.; Jourdie, V.; Jakab, G.; Karlen, D.; Held, M.; Mauch-Mani, B.; Turlings, T.C. Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 2007, 49, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Zhurov, V.; Navarro, M.; Martinez, M.; Cazaux, M.; Auger, P.; Migeon, A.; Santamaria, M.E.; Wybouw, N.; Diaz, I.; et al. Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Mol. Plant Microbe Interact. 2015, 28, 343–361. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wang, Y.; Ding, G.; Li, W.; Yang, G.; He, N. Biotic stress-induced expression of mulberry cystatins and identification of cystatin exhibiting stability to silkworm gut proteinases. Planta 2015, 242, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Campos, R.; Torres-Acosta, J.A.; Saucedo-Arias, L.J.; Gomez-Lim, M.A. The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 1999, 17, 1223–1226. [Google Scholar] [PubMed]
- Martinez, M.; Abraham, Z.; Gambardella, M.; Echaide, M.; Carbonero, P.; Diaz, I. The strawberry gene Cyf1 encodes a phytocystatin with antifungal properties. J. Exp. Bot. 2005, 56, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.M.; dos Reis, S.P.; de Souza, C.R. Phytocystatins and their potential to control plant diseases caused by fungi. Protein Pept. Lett. 2015, 22, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Lopez-Solanilla, E.; Rodriguez-Palenzuela, P.; Carbonero, P.; Diaz, I. Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol. Plant Microbe Interact. 2003, 16, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Kiggundu, A.; Goulet, M.C.; Goulet, C.; Dubuc, J.F.; Rivard, D.; Benchabane, M.; Pepin, G.; van der Vyver, C.; Kunert, K.; Michaud, D. Modulating the proteinase inhibitory profile of a plant cystatin by single mutations at positively selected amino acid sites. Plant J. 2006, 48, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Goulet, M.C.; Dallaire, C.; Vaillancourt, L.P.; Khalf, M.; Badri, A.M.; Preradov, A.; Duceppe, M.O.; Goulet, C.; Cloutier, C.; Michaud, D. Tailoring the specificity of a plant cystatin toward herbivorous insect digestive cysteine proteases by single mutations at positively selected amino acid sites. Plant Physiol. 2008, 146, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Terra, W.R.; Ferreira, C. Biochemistry and molecular biology of digestion. In Insect Molecular Biology and Biochemistry; Gilbert, L.I., Ed.; Elsevier: New York, NY, USA, 2012; pp. 365–418. [Google Scholar]
- Vinokurov, K.S.; Elpidina, E.N.; Oppert, B.; Prabhakar, S.; Zhuzhikov, D.P.; Dunaevsky, Y.E.; Belozersky, M.A. Diversity of digestive proteinases in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae. Comp. Biochem. Physiol. 2006, 145, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Bigham, M.; Hosseininaveh, V. Digestive proteolytic activity in the pistachio green stink bug, Brachynema germari Kolenati (Hemiptera: Pentatomidae). J. Asia Pac. Entomol. 2010, 13, 221–227. [Google Scholar] [CrossRef]
- Rispe, C.; Kutsukake, M.; Doublet, V.; Hudaverdian, S.; Legeai, F.; Simon, J.C.; Tagu, D.; Fukatsu, T. Large gene family expansion and variable selective pressures for cathepsin B in aphids. Mol. Biol. Evol. 2008, 25, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, L.; Martinez, M.; Alvarez-Alfageme, F.; Castañera, P.; Smagghe, G.; Diaz, I.; Ortego, F. A barley cysteine-protease inhibitor reduces the performance of two aphid species in artificial diets and transgenic arabidopsis plants. Transgenic Res. 2011, 20, 305–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrillo, L.; Martinez, M.; Ramessar, K.; Cambra, I.; Castañera, P.; Ortego, F.; Diaz, I. Expression of a barley cystatin gene in maize enhances resistance against phytophagous mites by altering their cysteine-proteases. Plant Cell Rep. 2011, 30, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santamaria, M.E.; Gonzalez-Cabrera, J.; Martinez, M.; Grbic, V.; Castañera, P.; Diaz, I.; Ortego, F. Digestive proteases in bodies and faeces of the two-spotted spider mite, Tetranychus urticae. J. Insect Physiol. 2015, 78, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shukle, R.; Mittapalli, O.; Zhu, Y.C.; Reese, J.C.; Wang, H.; Hua, B.Z.; Chen, M.S. The gut transcriptome of a gall midge, Mayetiola destructor. J. Insect Physiol. 2010, 56, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Pauchet, Y.; Wilkinson, P.; Vogel, H.; Nelson, D.R.; Reynolds, S.E.; Heckel, D.G.; ffrench-Constant, R.H. Pyrosequencing the Manduca sexta larval midgut transcriptome: Messages for digestion, detoxification and defence. Insect Mol. Biol. 2010, 19, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Vila, L.; Quilis, J.; Meynard, D.; Breitler, J.C.; Marfa, V.; Murillo, I.; Vassal, J.M.; Messeguer, J.; Guiderdoni, E.; SanSegundo, B. Expression of the maize proteinase inhibitor (Mpi) gene in rice plants enhances resistance against the striped stem borer (Chilo suppressalis): Effects on larval growth and insect gut proteinases. Plant Biotechnol. J. 2005, 3, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Chougule, N.P.; Giri, A.P.; Sainani, M.N.; Gupta, V.S. Gene expression patterns of Helicoverpa armigera gut proteases. Insect Biochem. Mol. Biol. 2005, 35, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R.; Clem, R.J. Insect Proteases. In Insect Molecular Biology and Biochemistry; Gilbert, L.I., Ed.; Elsevier: New York, NY, USA, 2012; pp. 346–364. [Google Scholar]
- Santamaria, M.E.; Hernandez-Crespo, P.; Ortego, F.; Grbic, V.; Grbic, M.; Diaz, I.; Martinez, M. Cysteine peptidases and their inhibitors in Tetranychus urticae: A comparative genomic approach. BMC Genom. 2012, 13, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koiwa, H.; Shade, R.E.; Zhu-Salzman, K.; D’Urzo, M.P.; Murdock, L.L.; Bressan, R.A.; Hasegawa, P.M. A plant defensive cystatin (soyacystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera). FEBS Lett. 2000, 471, 67–70. [Google Scholar] [CrossRef]
- Cristofoletti, P.T.; Ribeiro, A.F.; Terra, W.R. The cathepsin L-like proteinases from the midgut of Tenebrio molitor larvae: Sequence, properties, immunocytochemical localization and function. Insect Biochem. Mol. Biol. 2005, 35, 883–901. [Google Scholar] [CrossRef] [PubMed]
- Soares-Costa, A.; Dias, A.B.; Dellamano, M.; de Paula, F.F.P.; Carmona, A.K.; Terra, W.R.; Henrique-Silva, F. Digestive physiology and characterization of digestive cathepsin L-like proteinase from the sugarcane weevil Sphenophorus levis. J. Insect Physiol. 2011, 57, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.T.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaud, D.; Bernier-Vadnais, N.; Overney, S.; Yelle, S. Constitutive expression of digestive cysteine proteinase forms during development of the Colorado potato beetle, Leptinotarsa decemlineata Say (Coleoptera: Chrysomelidae). Insect Biochem. Mol. Biol. 1995, 25, 1041–1048. [Google Scholar] [CrossRef]
- Overney, S.; Fawe, A.; Yelle, S.; Michaud, D. Diet-related plasticity of the digestive proteolytic system in larvae of the Colorado potato beetle (Leptinotarsa decemlineata Say). Arch. Insect Biochem. Physiol. 1997, 36, 241–250. [Google Scholar] [CrossRef]
- Spit, J.; Badisco, L.; Verlinden, H.; van Wielendaele, P.; Zels, S.; Dillen, S.; Vanden-Broeck, J. Peptidergic control of food intake and digestion in insects. Can. J. Zool. 2012, 90, 489–506. [Google Scholar]
- Billingsley, P.F.; Lehane, M.J. Structure and ultrastructure of the insect midgut. In Biology of the Insect Midgut; Lehane, M.J., Billingsley, P.F., Eds.; Chapman and Hall: London, UK, 1996; pp. 3–30. [Google Scholar]
- Dutt, S.; Singh, V.K.; Marla, S.S.; Kumar, A. In silico analysis of sequential, structural and functional diversity of wheat cystatins and its implication in plant defense. Genom. Proteom. Bioinform. 2010, 8, 42–56. [Google Scholar] [CrossRef]
- Koiwa, H.; Shade, RE.; Zhu-Salzman, K.; Subramanian, L.; Murdock, L.L.; Nielsen, S.S.; Bressan, R.A.; Hasegawa, P.M. Phage display selection can differentiate insecticidal activity of soybean cystatins. Plant J. 1998, 14, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, H.; D’Urzo, M.P.; Assfalg-Machleidt, I.; Zhu-Salzman, K.; Shade, R.E.; An, H.; Murdock, L.L.; Machleidt, W.; Bressan, R.A.; Hasegawa, P.M. Phage display selection of hairpin loop soyacystatin variants that mediate high affinity inhibition of a cysteine proteinase. Plant J. 2001, 27, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, F.; Rhéaume, A.J.; Goulet, M.C.; Vorster, J.; Michaud, D. Discrimination of differentially inhibited cysteine proteases by activity-based profiling using cystatin variants with tailored specificities. J. Proteome Res. 2012, 11, 5983–5993. [Google Scholar] [CrossRef] [PubMed]
- Vorster, J.; Rasoolizadeh, A.; Goulet, M.C.; Cloutier, C.; Sainsbury, F.; Michaud, D. Positive selection of digestive Cys proteases in herbivorous Coleoptera. Insect Biochem. Mol. Biol. 2015, 65, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Alfageme, F.; Martınez, M.; Pascual-Ruiz, S.; Castañera, P.; Diaz, I.; Ortego, F. Effects of potato plants expressing a barley cystatin on the predatory bug Podisus maculiventris via herbivorous prey feeding on the plant. Transgenic Res. 2007, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Bonade-Bottino, M.; Pham-Delegue, M.H.; Jouanin, L. Two strains of cabbage seed weevil (Coleoptera: Curculionidae) exhibit differential susceptibility to a transgenic oilseed rape expressing oryzacystatin I. J. Insect Physiol. 1998, 44, 569–577. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Koiwa, H.; Salzman, R.A.; Shade, R.E.; Ahn, J.E. Cowpea bruchid Callosobruchus maculatus uses a three-component strategy to overcome a plant defensive cysteine protease inhibitor. Insect Mol. Biol. 2003, 12, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Rivard, D.; Cloutier, C.; Michaud, D. Colorado Potato Beetles show differential digestive compensatory responses to host plants expressing distinct sets of defense proteins. Arch. Insect Biochem. Physiol. 2004, 55, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Jouanin, L.; Pham-Delegue, M.; Bonade-Bottino, M.; Bartlet, E.; Zaccomer, B.; Le Metayer, M.; Girard, C. Growth simulation of beetle larvae reared on a transgenic oilseed rape expressing a cysteine proteinase inhibitor. J. Insect Physiol. 1998, 44, 263–270. [Google Scholar] [PubMed]
- Lara, P.; Ortego, F.; Gonzalez-Hidalg, E.; Castañera, P.; Carbonero, P.; Diaz, I. Adaptation of Spodoptera exigua (Lepidoptera: Noctuidae) to barley trypsin inhibitor BTI-CMe expressed in transgenic tobacco. Transgenic Res. 2000, 9, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Jongsma, M.A.; Bakker, P.L.; Peters, J.; Bosch, D.; Stiekema, W.J. Adaptation of Spodoptera exigua larvae to plant proteinase inhibitors by induction of gut proteinase activity insensitive to inhibition. Proc. Natl. Acad. Sci. USA 1995, 92, 8041–8045. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, C.; Jean, C.; Fournier, M.; Yelle, S.; Michaud, D. Adult Colorado potato beetles, Leptinotarsa decemlineata compensate for nutritional stress on oryzacystatin I-transgenic potato plants by hypertrophic behavior and over-production of insensitive proteases. Arch. Insect Biochem. Physiol. 2000, 44, 69–81. [Google Scholar] [CrossRef]
- Giri, A.P.; Harsulkar, A.M.; Deshpande, V.V.; Sainani, M.N.; Gupta, V.S.; Ranjekar, P.K. Chickpea defensive proteinase inhibitors can be inactivated by podborer gut proteinases. Plant Physiol. 1998, 116, 393–401. [Google Scholar] [CrossRef]
- Brunelle, F.; Cloutier, C.; Michaud, D. Colorado potato beetles compensate for tomato cathepsin D inhibitor expressed in transgenic potato. Arch. Insect Biochem. Physiol. 2004, 42, 88–98. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Zeng, R. Insect response to plant defensive protease inhibitors. Annu. Rev. Entomol. 2015, 60, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, C.; Fournier, M.; Jean, C.; Yelle, S.; Michaud, D. Growth compensation and faster development of Colorado potato beetle (Coleoptera: Chrysomelidae) feeding on potato foliage expressing oryzacystatin I. Arch. Insect Biochem. Physiol. 1999, 40, 69–79. [Google Scholar] [CrossRef]
- Santamaria, M.E.; Cambra, I.; Martinez, M.; Pozancos, C.; Gonzalez-Melendi, P.; Grbic, V.; Castañera, P.; Ortego, F.; Diaz, I. Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae. PLoS ONE 2012, 7, e43011. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, E.; Cloutier, C.; Michaud, D. Oryzacystatin I expressed in transgenic potato induces digestive compensation in an insect natural predator via its herbivorous prey feeding on the plant. Mol. Ecol. 2003, 12, 2439–2446. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, E.; Michaud, D.; Cloutier, C. Molecular interaction between an insect predator and its herbivore prey on transgenic potato expressing a cysteine proteinase inhibitor from rice. Mol. Ecol. 2003, 12, 2429–2437. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, F.C.S.; Silva, C.P.; Alexandre, D.; Samuels, R.I.; Soares, E.L.; Aragao, F.J.L.; Palmisano, G.; Domont, G.B.; Roepstoff, P.; Campos, F.A.P. Global proteome changes in larvae of Callosobruchus maculatus (Coleoptera: Chrysomelidae: Bruchinae) following ingestion of a cysteine proteinase inhibitor. Proteomics 2012, 12, 2704–2715. [Google Scholar] [CrossRef] [PubMed]
- Pillay, P.; Schluter, U.; van Wyck, S.G.; Kunert, K.J.; Vorster, B.J. Proteolysis of recombinant proteins in bioengineered plant cells. Bioengineered 2014, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pernas, M.; Sanchez-Monge, R.; Gomez, L.; Salcedo, G. A chesnut seed cystatin differentially effective against cysteine proteinases from closely related pests. Plant Mol. Biol. 1998, 38, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Rodriguez, S.; Galvan-Ramirez, J.P.; Guerrero-Rangel, A.; Cedro-Tanda, A. Multifunctional amaranth cystatin inhibits endogenous and digestive insect cysteine endopeptidases: A potential tool to prevent proteolysis and for the control of insect pests. Biotechnol. Appl. Biochem. 2015, 62, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Azzouz, H.; Cherqui, A.; Campan, E.D.; Rahbe, Y.; Duport, G.; Jouanin, L.; Kaiser, L.; Giordanengo, P. Effects of plant protease inhibitors, oryzacystatin I and soybean Bowman-Birk inhibitor, on the aphid Macrosiphum euphorbiae (Homoptera, Aphididae) and its parasitoid Aphelinus abdominalis (Hymenoptera, Aphelinidae). J. Insect Physiol. 2005, 51, 75–86. [Google Scholar] [CrossRef] [PubMed]
- James, C. 20th Anniversary (1996 to 2015) of the Global Commercialization of Biotech Crops and Biotech Crop Highlights in 2015. In ISAAA Brief; The International Service for the Acquisition of Agri-biotech Applications: Ithaca, NY, USA, 2015. [Google Scholar]
- Lawo, N.C.; Wackers, F.L.; Romeis, J. Indian Bt cotton in control of phytophagous insects. PLoS ONE 2009, 4, e4804. [Google Scholar]
- Li, Y.; Romeis, J. Bt maize expressing Cry3Bb1 does not harm the spider mite, Tetranychus urticae, or its ladybird beetle predator, Stethorus punctillum. BioControl 2010, 56, 157–164. [Google Scholar] [CrossRef]
- Esteves Filho, J.; Oliveira, V.; Torres, J.B.; Gondim, M.G.C. Biologia comparada e comportamento de Tetranychus urticae Koch (Acari: Tetranychidae) e Phytoseiulus macropilis (Banks) (Acari: Phytoseiidae) em algodoeiro BollgardTM e isolinha não-transgênica. Neotrop. Entomol. 2010, 39, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.C.; Yao, J.; Long, L.P.; Romeis, J.; Shelton, A.M. Bt crops benefit natural enemies to control non-target pests. Sci. Rep. 2015, 5, 16636. [Google Scholar] [CrossRef] [PubMed]
- Lecardonnel, A.; Chauvin, L.; Jouanin, L.; Beaujean, A.; Prevost, G.; Sangwan-Norreel, B. Effects of rice cystatin I expression in transgenic potato on Colorado potato beetle larvae. Plant Sci. 1999, 140, 71–79. [Google Scholar] [CrossRef]
- Cingel, A.; Savic, J.; Vinterhalter, B.; Vinterhalter, D.; Kostic, M.; Jovanovic, D.S.; Smigocki, A.; Ninkovic, S. Growth and development of Colorado potato beetle larvae, Leptinotarsa decemlineata, on potato plants expressing the oryzacystatin II proteinase inhibitor. Transgenic Res. 2015, 24, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Cingel, A.; Savic, J.; Cosic, T.; Zdravkovic-Korac, S.; Momcilovic, I.; Smigocki, A.; Ninkovic, S. Pyramiding rice cystatin OCI and OCII genes in transgenic potato (Solanum tuberosum L.) for resistance to Colorado potato beetle (Leptinotarsa decemlineata Say). Euphytica 2014, 198, 425–438. [Google Scholar] [CrossRef]
- Cingel, A.; Savic, J.; Lazarevic, J.; Cosic, T.; Raspor, M.; Smigocki, A.; Ninkovic, S. Co-expression of the proteinase inhibitors oryzacystatin I and oryzacystatin II in transgenic potato alters Colorado potato beetle larval development. Insect Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gatehouse, A.M.R.; Down, R.E.; Powell, K.S.; Sauvion, N.; Rahbe, Y.; Newell, C.A.; Merryweather, A.; Hamilton, W.D.O.; Gatehouse, J.A. Transgenic potato plants with enhanced resistance to the peach-potato aphid Myzus persicae. Entomol. Exp. Appl. 1996, 79, 295–307. [Google Scholar] [CrossRef]
- Schuler, T.H.; Denholm, I.; Jouanin, L.; Clark, S.J.; Clarak, A.J.; Poppy, G.M. Population-scale laboratory studies of the effect of transgenic plants on nontarget insects. Mol. Ecol. 2001, 10, 1845–1853. [Google Scholar] [CrossRef] [PubMed]
- Rahbe, Y.; Deraison, C.; Bonade-Bottino, M.; Girard, C.; Nardon, C.; Jouanin, L. Effects of the cysteine protease inhibitor oryzacystatin (IOC-I) on different aphids and reduced performance of Myzus persicae on OC-I expressing transgenic oilseed rape. Plant Sci. 2003, 164, 441–450. [Google Scholar] [CrossRef]
- Cowgill, S.D.; Danks, C.; Atkinson, H.J. Multitrophic interactions involving genetically modified potatoes, nontarget aphids, natural enemies and hyperparasitoids. Mol. Ecol. 2004, 13, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.P.O.; Pereira, E.J.C.; Galvan, T.L.; Picanzo, M.C.; Picoli, E.A.T.; da Silva, D.J.H.; Fari, M.G.; Otoni, W.C. Effect of eggplant transformed with oryzacystatin gene on Myzus persicae and Macrosiphom euphorbiae. J. Appl. Entomol. 2006, 130, 84–90. [Google Scholar] [CrossRef]
- Chen, P.J.; Senthilkumar, R.; Jane, W.N.; He, Y.; Tian, Z.; Yeh, K.W. Transplastomic Nicotiana benthamiana plants expressing multiple defence genes encoding protease inhibitors and chitinase display broad-spectrum resistance against insects, pathogens and abiotic stresses. Plant Biotechnol. J. 2014, 12, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Outchkourov, N.S.; de Kogel, W.J.; Schuurman-de Bruin, A.; Abrahamson, M.; Jongsma, M.A. Specific cysteine protease inhibitors act as deterrents of western flower thrips, Frankliniella occidentalis (Pergande), in transgenic potato. Plant Biotechnol. J. 2004, 2, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Outchkourov, N.S.; de Kogel, W.J.; Wiegers, G.L.; Abrahamson, M.; Jongsma, M.A. Engineered multidomain cysteine protease inhibitors yield resistance against western flower thrips (Frankliniella occidentalis) in greenhouse trials. Plant Biotechnol. J. 2004, 2, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Bonnade-Bottino, M.; Lerin, J.; Zaccomer, B.; Jouanin, L. Physiological adaptation explains the insensitivity of Baris coerulescens to transgenic oilseed rape expressing oryzacystatin I. Insect Biochem. Mol. Biol. 1999, 29, 131–138. [Google Scholar] [CrossRef]
- Leple, J.C.; Bonade-Bottino, M.; Augustin, S.; Pilate, G.; Le Tan, V.D.; Deplanque, A.; Cornu, D.; Jouanin, L. Toxicity to Chrysomela tremulae (Coleoptera: Chrysomelidae) of transgenic poplars expressing a cysteine proteinase inhibitor. Mol. Breed. 1995, 1, 319–328. [Google Scholar] [CrossRef]
- Delledonne, M.; Allegro, G.; Belenghi, B.; Balestrazzi, A.; Picco, F.; Levine, A.; Zelasco, S.; Calligari, P.; Confalonieri, M. Transformation of white poplar (Populus alba L.) with a novel Arabidopsis thaliana cysteine proteinase inhibitor and analysis of insect pest resistance. Mol. Breed. 2001, 7, 35–42. [Google Scholar] [CrossRef]
- Ninkovic, S.; Miljus-Dukic, J.; Radovic, S.; Maksimovic, V.; Lazarevic, J.; Vinterhalter, B.; Neskovic, M.; Smigocki, A. Phytodecta fornicata Brüggemann resistance mediated by oryzacystatin II proteinase inhibitor transgene. Plant Cell Tissue Organ Cult. 2007, 91, 289–294. [Google Scholar] [CrossRef]
- Irie, K.; Hosoyama, H.; Takecuchi, T.; Iwabuchi, K.; Watanabe, H.; Abe, M.; Abe, K.; Arai, S. Transgenic rice established to express corn cystatin exhibits strong inhibitory activity against insect gut proteinases. Plant Mol. Biol. 1996, 30, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Ferry, N.; Raemaekers, R.J.M.; Majerus, M.E.N.; Jouanin, L.; Port, G.; Gatehouse, J.A.; Gatehouse, A.M.R. Impact of oilseed rape expressing the insecticidal cysteine protease inhibitor oryzacystatin on the beneficial predator Harmonia axyridis (Multicoloured Asian ladybeetle). Mol. Ecol. 2003, 12, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, M.; Kuroda, M.; Yoza, K.I.; Nishizawa, K.; Teraishi, M.; Mizutani, N.; Ito, K.; Moriya, S. Heterologous expression of corn cystatin in soybean and effect on growth of the stink bug. Biosci. Biotechnol. Biochem. 2012, 76, 2142–2145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, M.; Zhang, X.; Luan, H.; Diao, S.; Tian, Y.; Su, X. Laboratory and field evaluation of the transgenic Populus alba × Populus glandulosa expressing double coleopteran-resistance genes. Tree Physiol. 2016, 31, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, R.; Cheng, C.P.; Yeh, K.W. Genetically pyramiding proteaseinhibitor genes for dual broad-spectrum resistance against insect and phytopathogens in transgenic tobacco. Plant Biotechnol. J. 2010, 8, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuween, T.; Demaeght, P.; Osborne, E.J.; Dermauw, W.; Gohlke, S.; Nauend, R.; Grbic, M.; Tirry, L.; Merzendorferc, H.; Clark, R.M. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. USA 2012, 105, 598–595. [Google Scholar] [CrossRef] [PubMed]



| Target Pest | Cystatin | Transgenic Plant | Reference | |
|---|---|---|---|---|
| Order | Species | |||
| Homoptera | Myzus persicae | OC-I | Potato | [88] |
| Oilseed rape | [89,90] | |||
| Eggplant | [92] | |||
| OC-I∆D86 | Potato | [91] | ||
| HvCPI-6 | Arabidopsis | [37] | ||
| Coleoptera | Baris coerulescens | OC-I | Oilseed rape | [96] |
| Ceutorhynchus assimilis | OC-I | Oilseed rape | [60,63] | |
| Chrysomela tremulae | OC-I | Poplar | [97] | |
| Chrysomela populi | AtCYS | Poplar | [98] | |
| Leptinotarsa decemlineata | OC-I | Potato | [66,72,73,74,84] | |
| OC-II | Potato | [85] | ||
| HvCPI-1 C→G | Potato | [59] | ||
| Phytodecta fornicata | OC-II | Alfalfa | [99] | |
| Psylliodes chrysocephala | OC-I | Oilseed rape | [63] | |
| Sitophilus zeamais | OC-I | Rice | [100] | |
| Lepidoptera | Plutella xylostella | OC-I | Oilseed rape | [101] |
| Spodoptera littoralis | HvCPI-1 C→G | Potato | [38] | |
| Hemiptera | Macrosiphum euphorbiae | OC-I | Eggplant | [92] |
| OC-I∆D86 | Potato | [91] | ||
| Riptortus clavatus | CCI | Soybean | [102] | |
| Acarina | Brevipalpuls chilensis | HvCPI-1 C→G | Potato | [38] |
| Tetranychus urticae | HvCPI-6 | Maize | [38] | |
| HvCPI-6 | Arabidopsis | [39,71] | ||
| Target Pest | Cystatin + Proteins | Transgenic Plant | Reference | |
|---|---|---|---|---|
| Order | Species | |||
| Coleoptera | Leptinotarsa decemlineata | OCI + OCII | Potato | [86,87] |
| OC-I∆D86 + CpTI | Arabidopsis | [62] | ||
| Plagiodera versicolora | OC-I + CRY3A | Poplar | [103] | |
| Lepidoptera | Helicoverpa armigera | CeCPI + Sporamin | Tobacco | [104] |
| Spodoptera exigua | CeCPI + Sporamin + chitinase | Tobacco | [93] | |
| Spodoptera littura | CeCPI + Sporamin + chitinase | Tobacco | [93] | |
| Thysanoptera | Frankiniella occidentalis | Engineered PCM + domains | Potato | [95] |
| Acarina | Tetranychus urticae | HvCPI-6 + CMe | Arabidopsis | [70] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez, M.; Santamaria, M.E.; Diaz-Mendoza, M.; Arnaiz, A.; Carrillo, L.; Ortego, F.; Diaz, I. Phytocystatins: Defense Proteins against Phytophagous Insects and Acari. Int. J. Mol. Sci. 2016, 17, 1747. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101747
Martinez M, Santamaria ME, Diaz-Mendoza M, Arnaiz A, Carrillo L, Ortego F, Diaz I. Phytocystatins: Defense Proteins against Phytophagous Insects and Acari. International Journal of Molecular Sciences. 2016; 17(10):1747. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101747
Chicago/Turabian StyleMartinez, Manuel, Maria Estrella Santamaria, Mercedes Diaz-Mendoza, Ana Arnaiz, Laura Carrillo, Felix Ortego, and Isabel Diaz. 2016. "Phytocystatins: Defense Proteins against Phytophagous Insects and Acari" International Journal of Molecular Sciences 17, no. 10: 1747. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101747