Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions
Abstract
:1. Introduction
2. Bacterial Carotenoid Oxygenases
2.1. Structural Studies
2.2. Substrate Specificity
2.2.1. Apocarotenoid Cleavage Oxygenases (ACO)
2.2.2. CCDs with Symmetrical Mode of Action
2.2.3. Carotenoid Oxygenases Cleaving at Different Positions
3. Fungal Carotenoid Oxygenases
3.1. Fungal CCDs Involved in Retinal Production
3.2. Fungal CCDs Involved in Trisporic Acid Production
3.3. Fungal CCDs Involved in Neurosporaxanthin Production
4. Plant CCDs
4.1. CCD1 and CCD2 Subfamilies
4.1.1. Mode of Action and Functional Implications
4.1.2. CCD1 Gene Family and Genomic Organization
4.1.3. Mode of Action of CCD2
4.1.4. Structure of CCD1 and CCD2 Enzymes
4.1.5. Regulation of CCD1 and CCD2 Enzymes
4.2. The CCD4 Subfamily
4.2.1. Enzyme Structure and Functional Implications
4.2.2. Gene Family and Genomic Organization
4.2.3. CCD4 Activity: Carotenoid Substrates and Cleavage Products
4.2.4. Expression Pattern in Plant Tissues
4.3. The CCD7 and CCD8 Subfamilies
5. CCDs in Algae
6. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. Biosynthesis and Metabolism; Birkhäuser Verlag: Basel, Switzerland, 1998. [Google Scholar]
- Avalos, J.; Díaz-Sánchez, V.; García-Martínez, J.; Castrillo, M.; Ruger-Herreros, M.; Limón, M.C. Carotenoids. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.F., García-Estrada, C., Zeilinger, S., Eds.; Springer Science-Business Media: New York, NY, USA, 2014; pp. 149–185. [Google Scholar]
- Cazzonelli, C.I. Carotenoids in nature: Insights from plants and beyond. Funct. Plant Biol. 2011, 38, 833–847. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Jarvik, T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 2010, 328, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G. Genetics of eubacterial carotenoid biosynthesis: A colorful tale. Annu. Rev. Microbiol. 1997, 51, 629–659. [Google Scholar] [CrossRef] [PubMed]
- Mirkovic, T.; Ostroumov, E.E.; Anna, J.M.; van Grondelle, R.; Shoeles, G.; Scholes, G.D. Light absorption and energy transfer in the antenna complexes of photosynthetic organisms. Chem. Rev. 2016. [Google Scholar] [CrossRef] [PubMed]
- Britton, G. Functions of carotenoid metabolites and breakdown products. In Carotenoids; Britton, G., Liaaen-Jensen, S., Pfander, H., Eds.; Birkhäuser: Basel, Switzerland, 2008. [Google Scholar]
- Marasco, E.K.; Schmidt-Dannert, C. Identification of bacterial carotenoid cleavage dioxygenase homologues that cleave the interphenyl α,β-double bond of stilbene derivatives via a monooxygenase reaction. Chembiochem 2008, 9, 1450–1461. [Google Scholar] [CrossRef] [PubMed]
- Goodman, D.S.; Huang, H.S. Biosynthesis of vitamin A with rat intestinal enzymes. Science 1965, 149, 879–880. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Priya, R.; Siva, R. Analysis of phylogenetic and functional diverge in plant nine-cis-epoxycarotenoid dioxygenase gene family. J. Plant Res. 2015, 128, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R. Abscisic acid synthesis and response. Arabidopsis Book 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Kanno, Y.; Frey, A.; North, H.M.; Marion-Poll, A. Dissection of Arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 2016, 246, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that aba synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis-epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Jüttner, F.; Höflacher, B. Evidence of β-carotene 7,8(7′,8′) oxygenase (β-cyclocitral, crocetindial generating) in Microcystis. Arch. Microbiol. 1985, 141, 337–343. [Google Scholar] [CrossRef]
- Balashov, S.P.; Lanyi, J.K. Xanthorhodopsin: Proton pump with a carotenoid antenna. Cell. Mol. Life Sci. 2007, 64, 2323–2328. [Google Scholar] [CrossRef] [PubMed]
- Grote, M.; O’Malley, M.A. Enlightening the life sciences: The history of halobacterial and microbial rhodopsin research. FEMS Microbiol. Rev. 2011, 35, 1082–1099. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-H.; Trivedi, V.D.; Spudich, J.L. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 2003, 47, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Kloer, D.P.; Ruch, S.; Al-Babili, S.; Beyer, P.; Schulz, G.E. The structure of a retinal-forming carotenoid oxygenase. Science 2005, 308, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Kiser, P.D.; Lintig, J.V.; Palczewski, K. Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 2013, 539, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, E.; Gentleman, S.; Cunningham, F.X.; Miller-Ihli, N.J.; Redmond, T.M. Key role of conserved histidines in recombinant mouse β-carotene 15,15′-monooxygenase-1 activity. J. Biol. Chem. 2005, 280, 29217–29223. [Google Scholar] [CrossRef] [PubMed]
- Redmond, T.M.; Poliakov, E.; Yu, S.; Tsai, J.-Y.; Lu, Z.; Gentleman, S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. USA 2005, 102, 13658–13663. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Moiseyev, G.; Chen, Y.; Ma, J.-X. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 2005, 579, 5414–5418. [Google Scholar] [CrossRef] [PubMed]
- Kloer, D.P.; Schulz, G.E. Structural and biological aspects of carotenoid cleavage. Cell. Mol. Life Sci. 2006, 63, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Ruch, S.; Beyer, P.; Ernst, H.; Al-Babili, S. Retinal biosynthesis in eubacteria: In vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Mol. Microbiol. 2005, 55, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Scherzinger, D.; Al-Babili, S. In vitro characterization of a carotenoid cleavage dioxygenase from Nostoc sp. PCC 7120 reveals a novel cleavage pattern, cytosolic localization and induction by highlight. Mol. Microbiol. 2008, 69, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Kim, S.H.; Lee, P.C. New insight into the cleavage reaction of Nostoc sp. strain PCC 7120 carotenoid cleavage dioxygenase in natural and nonnatural carotenoids. Appl. Environ. Microbiol. 2013, 79, 3336–3345. [Google Scholar] [CrossRef] [PubMed]
- Scherzinger, D.; Ruch, S.; Kloer, D.P.; Wilde, A.; Al-Babili, S. Retinal is formed from apo-carotenoids in Nostoc sp. PCC7120: In vitro characterization of an apo-carotenoid oxygenase. Biochem. J. 2006, 398, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Marasco, E.K.; Vay, K.; Schmidt-Dannert, C. Identification of carotenoid cleavage dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. J. Biol. Chem. 2006, 281, 31583–31593. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Bustamante, E.; Sánchez, S. Microbial production of C13-norisoprenoids and other aroma compounds via carotenoid cleavage. Crit. Rev. Microbiol. 2007, 33, 211–230. [Google Scholar] [CrossRef] [PubMed]
- Höckelmann, C.; Jüttner, F. Off-flavours in water: Hydroxyketones and β-ionone derivatives as new odor compounds of freshwater cyanobacteria. Flavour. Fragr. J. 2005, 20, 387–394. [Google Scholar] [CrossRef]
- Scherzinger, D.; Scheffer, E.; Bär, C.; Ernst, H.; Al-Babili, S. The Mycobacterium tuberculosis ORF Rv0654 encodes a carotenoid oxygenase mediating central and excentric cleavage of conventional and aromatic carotenoids. FEBS J. 2010, 277, 4662–4673. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Seo, E.-S.; Oh, D.-K. Characterization of an apo-carotenoid 13,14-dioxygenase from Novosphingobium aromaticivorans that converts β-apo-80-carotenal to β-apo-13-carotenone. Biotechnol. Lett. 2012, 34, 1851–1856. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.; Bóna-Lovász, J.; Beuttler, H.; Altenbuchner, J. In vivo and in vitro studies on the carotenoid cleavage oxygenases from Sphingopyxis alaskensis RB2256 and Plesiocystis pacifica SIR-1 revealed their substrate specificities and non-retinal-forming cleavage activities. FEBS J. 2012, 279, 3911–3924. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.; Cerdá-Olmedo, E. Fungal carotenoid production. In Handbook of Fungal Biotechnology, 2nd ed.; Arora, D.K., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 2004. [Google Scholar]
- Sandmann, G.; Misawa, N. Fungal carotenoids. In Industrial Applications; Osiewacz, H.D., Ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Avalos, J.; Mackenzie, A.; Nelki, D.S.; Bramley, P.M. Terpenoid biosynthesis in cell-extracts of wild type and mutant strains of Gibberella fujikuroi. Biochim. Biophys. Acta 1988, 966, 257–265. [Google Scholar] [CrossRef]
- Salgado, L.M.; Avalos, J.; Bejarano, E.R.; Cerdá-Olmedo, E. Correlation between in vivo and in vitro carotenogenesis in Phycomyces. Phytochemistry 1991, 30, 2587–2591. [Google Scholar] [CrossRef]
- Brown, L.S. Fungal rhodopsins and opsin-related proteins: Eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 2004, 3, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Bieszke, J.A.; Braun, E.L.; Bean, L.E.; Kang, S.; Natvig, D.O.; Borkovich, K.A. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA 1999, 96, 8034–8039. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Howlett, B.J. Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome 2001, 44, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Waschuk, S.A.; Bezerra, A.G., Jr.; Shi, L.; Brown, L.S. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl. Acad. Sci. USA 2005, 102, 6879–6883. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.M.; Prado-Cabrero, A.; Fernández-Martín, R.; Avalos, J. A gene of the opsin family in the carotenoid gene cluster of Fusarium fujikuroi. Curr. Gen. 2004, 46, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.F.; Avalos, J. Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J. Mol. Biol. 2009, 387, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Bieszke, J.A.; Spudich, E.N.; Scott, K.L.; Borkovich, K.A.; Spudich, J.L. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 1999, 38, 14138–14145. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, J.; Brunk, M.; Avalos, J.; Terpitz, U. The CarO rhodopsin of the fungus Fusarium fujikuroi is a light-driven proton pump that retards spore germination. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Wyss, A. Carotene oxygenases: A new family of double bond cleavage enzymes. J. Nutr. 2004, 134, 246S–250S. [Google Scholar] [PubMed]
- Thewes, S.; Prado-Cabrero, A.; Prado, M.M.; Tudzynski, B.; Avalos, J. Characterization of a gene in the car cluster of Fusarium fujikuroi that codes for a protein of the carotenoid oxygenase family. Mol. Genet. Genom. 2005, 274, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Prado-Cabrero, A.; Scherzinger, D.; Avalos, J.; Al-Babili, S. Retinal biosynthesis in fungi: Characterization of the carotenoid oxygenase CarX from Fusarium fujikuroi. Eukaryot. Cell 2007, 6, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Sánchez, V.; Estrada, A.F.; Limón, M.C.; Al-Babili, S.; Avalos, J. The oxygenase CAO-1 of Neurospora crassa is a resveratrol cleavage enzyme. Eukaryot. Cell 2013, 12, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.F.; Brefort, T.; Mengel, C.; Díaz-Sánchez, V.; Alder, A.; Al-Babili, S.; Avalos, J. Ustilago maydis accumulates β-carotene at levels determined by a retinal-forming carotenoid oxygenase. Fungal Genet. Biol. 2009, 46, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Austin, D.G.; Bu’Lock, J.D.; Winstanley, D.J. Trisporic acid biosynthesis and carotenogenesis in Blakesleea trispora. Biochem. J. 1969, 113. [Google Scholar] [CrossRef]
- Austin, D.J.; Bu’Lock, J.D.; Drake, D. The biosynthesis of trisporic acids from β-carotene via retinal and trisporol. Experientia 1970, 26, 348–349. [Google Scholar] [CrossRef] [PubMed]
- Caglioti, L.; Cainelli, G.; Camerino, B.; Mondelli, R.; Prieto, A.; Quilico, A.; Salvatori, T.; Selva, A. The structure of trisporic-c acid. Tetrahedron 1966, 22, 175–187. [Google Scholar] [CrossRef]
- Sutter, R.P.; Dadok, J.; Bothner-By, A.A.; Smith, R.R.; Mishra, P.K. Cultures of separated mating types of Blakeslea trispora make D and E forms of trisporic acids. Biochemistry 1989, 28, 4060–4066. [Google Scholar] [CrossRef] [PubMed]
- Barrero, A.F.; Herrador, M.M.; Arteaga, P.; Gil, J.; González, J.A.; Alcalde, E.; Cerdá-Olmedo, E. New apocarotenoids and β-carotene cleavage in Blakeslea trispora. Org. Biomol. Chem. 2011, 9, 7190–7195. [Google Scholar] [CrossRef] [PubMed]
- Polaino, S.; Gonzalez-Delgado, J.A.; Arteaga, P.; Herrador, M.M.; Barrero, A.F.; Cerdá-Olmedo, E. Apocarotenoids in the sexual interaction of Phycomyces blakesleeanus. Org. Biomol. Chem. 2012, 10, 3002–3009. [Google Scholar] [CrossRef] [PubMed]
- Burmester, A.; Richter, M.; Schultze, K.; Voelz, K.; Schachtschabel, D.; Boland, W.; Wöstemeyer, J.; Schimek, C. Cleavage of β-carotene as the first step in sexual hormone synthesis in zygomycetes is mediated by a trisporic acid regulated β-carotene oxygenase. Fungal Genet. Biol. 2007, 44, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Polaino, S.; Herrador, M.M.; Cerdá-Olmedo, E.; Barrero, A.F. Splitting of β-carotene in the sexual interaction of Phycomyces. Org. Biomol. Chem. 2010, 8, 4229–4231. [Google Scholar] [CrossRef] [PubMed]
- Tagua, V.G.; Medina, H.R.; Martín-Domínguez, R.; Eslava, A.P.; Corrochano, L.M.; Cerdá-Olmedo, E.; Idnurm, A. A gene for carotene cleavage required for pheromone biosynthesis and carotene regulation in the fungus Phycomyces blakesleeanus. Fungal Genet. Biol. 2012, 49, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Medina, H.R.; Cerdá-Olmedo, E.; Al-Babili, S. Cleavage oxygenases for the biosynthesis of trisporoids and other apocarotenoids in Phycomyces. Mol. Microbiol. 2011, 82, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, E.R.; Parra, F.; Murillo, F.J.; Cerdá-Olmedo, E. End-product regulation of carotenogenesis in Phycomyces. Arch. Microbiol. 1988, 150, 209–214. [Google Scholar] [CrossRef]
- Almeida, E.R.A.; Cerdá-Olmedo, E. Gene expression in the regulation of carotene biosynthesis in Phycomyces. Curr. Genet. 2008, 53, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Salgado, L.M.; Cerdá-Olmedo, E. Genetic interactions in the regulation of carotenogenesis in Phycomyces. Curr. Gen. 1992, 21, 67–71. [Google Scholar] [CrossRef]
- Czempinski, K.; Kruft, V.; Wöstemeyer, J.; Burmester, A. 4-dihydromethyltrisporate dehydrogenase from Mucor mucedo, an enzyme of the sexual hormone pathway: Purification, and cloning of the corresponding gene. Microbiology 1996, 142, 2647–2654. [Google Scholar] [CrossRef] [PubMed]
- Schimek, C.; Petzold, A.; Schultze, K.; Wetzel, J.; Wolschendorf, F.; Burmester, A.; Wöstemeyer, J. 4-dihydromethyltrisporate dehydrogenase, an enzyme of the sex hormone pathway in Mucor mucedo, is constitutively transcribed but its activity is differently regulated in (+) and (−) mating types. Fungal Genet. Biol. 2005, 42, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, J.; Scheibner, O.; Burmester, A.; Schimek, C.; Wöstemeyer, J. 4-dihydrotrisporin-dehydrogenase, an enzyme of the sex hormone pathway of Mucor mucedo: Purification, cloning of the corresponding gene, and developmental expression. Eukaryot. Cell 2009, 8, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Prado-Cabrero, A.; Estrada, A.F.; Al-Babili, S.; Avalos, J. Identification and biochemical characterization of a novel carotenoid oxygenase: Elucidation of the cleavage step in the Fusarium carotenoid pathway. Mol. Microbiol. 2007, 64, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-M.; Lee, J.; Lee, Y.-W. Characterization of carotenoid biosynthetic genes in the ascomycete Gibberella zeae. FEMS Microbiol. Lett. 2010, 302, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Saelices, L.; Youssar, L.; Holdermann, I.; Al-Babili, S.; Avalos, J. Identification of the gene responsible for torulene cleavage in the Neurospora carotenoid pathway. Mol. Genet. Genom. 2007, 278, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.; Corrochano, L.M. Carotenoid biosynthesis in Neurospora. In Neurospora: Genomics and Molecular Biology; Kasbekar, D.P., McCluskey, K., Eds.; Caister Academic Press: Wareham, UK, 2013. [Google Scholar]
- Avalos, J.; Estrada, A.F. Regulation by light in Fusarium. Fungal Genet. Biol. 2010, 47, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.F.; Youssar, L.; Scherzinger, D.; Al-Babili, S.; Avalos, J. The ylo-1 gene encodes an aldehyde dehydrogenase responsible for the last reaction in the Neurospora carotenoid pathway. Mol. Microbiol. 2008, 69, 1207–1220. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Sánchez, V.; Estrada, A.F.; Trautmann, D.; Al-Babili, S.; Avalos, J. The gene carD encodes the aldehyde dehydrogenase responsible for neurosporaxanthin biosynthesis in Fusarium fujikuroi. FEBS J. 2011, 278, 3164–3176. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Floss, D.S.; Strack, D. Apocarotenoids: Hormones, mycorrhizal metabolites and aroma volatiles. Planta 2010, 232, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 2001, 276, 25208–25211. [Google Scholar] [CrossRef] [PubMed]
- García-Limones, C.; Schnabele, K.; Blanco-Portales, R.; Luz Bellido, M.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J. Functional characterization of FaCCD1: A carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J. Agric. Food Chem. 2008, 56, 9277–9285. [Google Scholar] [CrossRef] [PubMed]
- Ibdah, M.; Azulay, Y.; Portnoy, V.; Wasserman, B.; Bar, E.; Meir, A.; Burger, Y.; Hirschberg, J.; Schaffer, A.A.; Katzir, N.; et al. Functional characterization of CmCCD1, a carotenoid cleavage dioxygenase from melon. Phytochemistry 2006, 67, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Ilg, A.; Beyer, P.; Al-Babili, S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 2009, 276, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, S.; Terrier, N.; Procureur, J.; Bigey, F.; Gunata, Z. A carotenoid cleavage dioxygenase from Vitis vinifera l: Functional characterization and expression during grape berry development in relation to C13-norisoprenoid accumulation. J. Exp. Bot. 2005, 56, 2721–2731. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Baldermann, S.; Kato, M.; Kurosawa, M.; Kurobayashi, Y.; Fujita, A.; Fleischmann, P.; Watanabe, N. Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans lour. J. Exp. Bot. 2010, 61, 2967–2977. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.; Rambla, J.L.; Santaella, M.; Gómez, M.D.; Orzaez, D.; Granell, A.; Gómez-Gómez, L. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in β-ionone release. J. Biol. Chem. 2008, 283, 24816–24825. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Schwartz, S.H.; Auldridge, M.; Taylor, M.G.; Klee, H.J. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles beta-ionone, pseudoionone, and geranylacetone. Plant J. 2004, 40, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Ji, J.; Wang, G.; Jin, C.; Guan, C.; Wu, G. Molecular cloning and characterization of a novel carotenoid cleavage dioxygenase 1 from Lycium chinense. Biotechnol. Appl. Biochem. 2014, 62, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Tan, B.C.; McCarty, D.R.; Klee, H.J. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 2008, 283, 11364–11373. [Google Scholar] [CrossRef] [PubMed]
- Ilg, A.; Bruno, M.; Beyer, P.; Al-Babili, S. Tomato carotenoid cleavage dioxygenases 1A and 1B: Relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Bio 2014, 4, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Schliemann, W.; Schmidt, J.; Strack, D.; Walter, M.H. RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol. 2008, 148, 1267–1282. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Matsumoto, H.; Ikoma, Y.; Okuda, H.; Yano, M. The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J. Exp. Bot. 2006, 57, 2153–2164. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Molnar, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef] [PubMed]
- Ilg, A.; Yu, Q.; Schaub, P.; Beyer, P.; Al-Babili, S. Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in golden rice endosperm suggests apocarotenoids as substrates in planta. Planta 2010, 232, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Lashbrooke, J.G.; Young, P.R.; Dockrall, S.J.; Vasanth, K.; Vivier, M.A. Functional characterisation of three members of the Vitis vinifera l. Carotenoid cleavage dioxygenase gene family. BMC Plant Biol. 2013, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Wooding, S. Olfaction: It makes a world of scents. Curr. Biol. 2013, 23, R677–R679. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, S.; Cin, V.D.; Fei, Z.; Li, H.; Bliss, P.; Taylor, M.G.; Klee, H.J.; Tieman, D.M. Flavour compounds in tomato fruits: Identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 2009, 60, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Suire, C.; Mutterer, J.; Camara, B. Oxidative remodeling of chromoplast carotenoids: Identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 2003, 15, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Walter, M.H. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signal. Behav. 2009, 4, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Yoneyama, K.; Hugill, C.; Quittenden, L.J.; Reid, J.B. Strigolactones: Internal and external signals in plant symbioses? Plant Signal. Behav. 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H. Role of carotenoid metabolism in the arbuscular mycorrhizal symbiosis. In Molecular Microbial Ecology of the Rhizosphere; Bruijn, F.J.D., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013. [Google Scholar]
- López-Raez, J.A.; Fernández, I.; García, J.M.; Berrio, E.; Bonfante, P.; Walter, M.H.; Pozo, M.J. Differential spatio-temporal expression of carotenoid cleavage dioxygenases regulates apocarotenoid fluxes during AM symbiosis. Plant Sci. 2015, 230, 59–69. [Google Scholar] [CrossRef] [PubMed]
- González-Jorge, S.; Ha, S.H.; Magallanes-Lundback, M.; Gilliland, L.U.; Zhou, A.; Lipka, A.E.; Nguyen, Y.N.; Angelovici, R.; Lin, H.; Cepela, J.; et al. CAROTENOID CLEAVAGE DIOXYGENASE4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 2013, 25, 4812–4826. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Isner, J.C.; Dogbo, O.; Camara, B. Oxidative tailoring of carotenoids: A prospect towards novel functions in plants. Trends Plant Sci. 2005, 10, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Hannoufa, A.; Soroka, J.; Xu, N.; Li, X.; Zebarjadi, A.; Gruber, M. Enhanced β-ionone emission in Arabidopsis over-expressing AtCCD1 reduces feeding damage in vivo by the crucifer flea beetle. Environ. Entomol. 2011, 40, 1622–1630. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Avila, N.L.; Narvaez-Zapata, J.A.; Ramírez-Benítez, J.E.; Aguilar-Espinosa, M.L.; Rivera-Madrid, R. Identification and expression pattern of a new carotenoid cleavage dioxygenase gene member from Bixa orellana. J. Exp. Bot. 2011, 62, 5385–5395. [Google Scholar] [CrossRef] [PubMed]
- González-Verdejo, C.I.; Obrero, A.; Roman, B.; Gómez, P. Expression profile of carotenoid cleavage dioxygenase genes in summer squash (Cucurbita pepo l.). Plant Foods Hum. Nutr. 2015, 70, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Imai, A.; Takahashi, S.; Nakayama, K.; Satoh, H. The promoter of the carotenoid cleavage dioxygenase 4a-5 gene of Chrysanthemum morifolium (CmCCD4a-5) drives petal-specific transcription of a conjugated gene in the developing flower. J. Plant Physiol. 2013, 170, 1295–1299. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jeong, J.C.; Park, S.; Bae, J.Y.; Ahn, M.J.; Lee, H.S.; Kwak, S.S. Down-regulation of sweetpotato lycopene β-cyclase gene enhances tolerance to abiotic stress in transgenic calli. Mol. Biol. Rep. 2014, 41, 8137–8148. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Hans, J.; Walter, M.H.; Matusova, R.; Beekwilder, J.; Verstappen, F.W.; Ming, Z.; van Echtelt, E.; Strack, D.; Bisseling, T.; et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 2008, 228, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Trapero, A.; Gómez, M.D.; Rubio-Moraga, A.; Gómez-Gómez, L. Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: A deeper study in Crocus sativus and its allies. Genomics 2010, 96, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Márquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [PubMed]
- Frusciante, S.; Diretto, G.; Bruno, M.; Ferrante, P.; Pietrella, M.; Prado-Cabrero, A.; Rubio-Moraga, A.; Beyer, P.; Gómez-Gómez, L.; Al-Babili, S.; et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 12246–12251. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gómez-Gómez, L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Nebauer, S.G.; Molina, R.V.; Gómez-Gómez, L. Saffron: Its phytochemistry, developmental processes, and biotechnological prospects. J. Agric. Food Chem. 2015, 63, 8751–8764. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Moraga, A.; Ahrazem, O.; Rambla, J.L.; Granell, A.; Gómez-Gómez, L. Crocins with high levels of sugar conjugation contribute to the yellow colors of early-spring flowering crocus tepals. PLoS ONE 2013, 8, e71946. [Google Scholar] [CrossRef] [PubMed]
- Jüttner, F.; Watson, S.B.; von Elert, E.; Koster, O. β-cyclocitral, a grazer defence signal unique to the cyanobacterium Microcystis. J. Chem. Ecol. 2010, 36, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Messing, S.A.; Gabelli, S.B.; Echeverria, I.; Vogel, J.T.; Guan, J.C.; Tan, B.C.; Klee, H.J.; McCarty, D.R.; Amzel, L.M. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 2010, 22, 2970–2980. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Argandoña, J.; Castillo-López, R.; Gómez-Gómez, L. Intron retention and rhythmic diel pattern regulation of carotenoid cleavage dioxygenase 2 during crocetin biosynthesis in saffron. Plant Mol. Biol. 2016, 91, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Moraga, A.R.; Nohales, P.F.; Pérez, J.A.F.; Gómez-Gómez, L. Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta 2004, 219, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Zhu, C.; Kuntz, M.; Sandmann, G. Light-dark regulation of carotenoid biosynthesis in pepper (Capsicum annuum) leaves. J. Plant Physiol. 2003, 160, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Labouré, A.-M.; Kuntz, M.; Sandmann, G. Comparison of carotenoid content, gene expression and enzyme levels in tomato (Lycopersicon esculentum) leaves. Z. Naturforschung C 2003, 58, 371–380. [Google Scholar] [CrossRef]
- Kato, M.; Ikoma, Y.; Matsumoto, H.; Sugiura, M.; Hyodo, H.; Yano, M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 2004, 134, 824–837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, G.; Kato, M.; Yamawaki, K.; Takagi, T.; Kiriiwa, Y.; Ikoma, Y.; Matsumoto, H.; Yoshioka, T.; Nesumi, H. Regulation of carotenoid accumulation and the expression of carotenoid metabolic genes in citrus juice sacs in vitro. J. Exp. Bot. 2011, 63, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Mockler, T.C. Unproductive alternative splicing and nonsense mRNAs: A widespread phenomenon among plant circadian clock genes. Biol. Direct 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Moraga, A.; Rambla, J.L.; Fernández-de-Carmen, A.; Trapero-Mozos, A.; Ahrazem, O.; Orzaez, D.; Granell, A.; Gómez-Gómez, L. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol. Biol. 2014, 85, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Zhang, L.; Matsuta, A.; Matsutani, K.; Yamawaki, K.; Yahata, M.; Wahyudi, A.; Motohashi, R.; Kato, M. Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 2013, 163, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 2009, 26, 351–358. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Alquézar, B.; Alós, E.; Medina, V.; Carmona, L.; Bruno, M.; Al-Babili, S.; Zacarías, L. A novel carotenoid cleavage activity involved in the biosynthesis of citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 2013, 64, 4461–4478. [Google Scholar] [CrossRef] [PubMed]
- Lätari, K.; Wüst, F.; Hübner, M.; Schaub, P.; Kim, G.B.; Matsubara, S.; Beyer, P.; Welsch, R. Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol. 2015, 168, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Vázquez, A.-O.; Cordoba, E.; Llamas, E.; San Román, C.; Nisar, N.; De la Torre, S.; Ramos-Vega, M.; de la Luz Gutiérrez-Nava, M.; Cazzonelli, C.I.; Pogson, B.J. An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 2014, 26, 2524–2537. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Ytterberg, A.J.; Peltier, J.-B.; van Wijk, K.J. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006, 140, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Brehelin, C.; Kessler, F.; van Wijk, K.J. Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 2007, 12, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Vidi, P.A.; Kessler, F.; Brehelin, C. Plastoglobules: A new address for targeting recombinant proteins in the chloroplast. BMC Biotechnol. 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, P.K.; Poliakov, A.; Bhuiyan, N.H.; Zybailov, B.; Sun, Q.; van Wijk, K.J. The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 2012, 158, 1172–1192. [Google Scholar] [CrossRef] [PubMed]
- Besagni, C.; Kessler, F. A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation. Planta 2013, 237, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Rottet, S.; Besagni, C.; Kessler, F. The role of plastoglobules in thylakoid lipid remodeling during plant development. Biochim. Biophys. Acta 2015, 1847, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, P.K.; Poliakov, A.; Giacomelli, L.; Friso, G.; Appel, M.; McQuinn, R.P.; Krasnoff, S.B.; Rowland, E.; Ponnala, L.; Sun, Q.; et al. Loss of plastoglobule kinases ABC1K1 and ABD1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 2013, 25, 1818–1839. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, R.; Bradbury, L.M.; Wurtzel, E.T. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 2010, 504, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Ono, M.; Osaki, M.; Konishi, I.; Sakaguchi, S. Differential effects of inhibition of bone morphogenic protein (BMP) signalling on T-cell activation and differentiation. Eur. J. Immunol. 2012, 42, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xie, Z.; Zhu, K.; Xu, Q.; Deng, X.; Pan, Z. Isolation and characterization of carotenoid cleavage dioxygenase 4 genes from different citrus species. Mol. Genet. Genom. 2015, 290, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Moraga, A.R.; Rambla, J.L.; Ahrazem, O.; Granell, A.; Gómez-Gómez, L. Metabolite and target transcript analyses during Crocus sativus stigma development. Phytochemistry 2009, 70, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wan, H.; Wu, Z.; Wang, R.; Ruan, M.; Ye, Q.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y. A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum lycopersicum. Plant Mol. Biol. Rep. 2015, 34, 512–543. [Google Scholar] [CrossRef]
- Priya, R.; Siva, R. Phylogenetic analysis and evolutionary studies of plant carotenoid cleavage dioxygenase gene. Gene 2014, 548, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.; Ducreux, L.J.; Morris, W.L.; Morris, J.A.; Suttle, J.C.; Ramsay, G.; Bryan, G.J.; Hedley, P.E.; Taylor, M.A. The metabolic and developmental roles of carotenoid cleavage dioxygenase 4 from potato. Plant Physiol. 2010, 154, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Butchko, R.A.E.; Brown, D.W.; Busman, M.; Tudzynski, B.; Wiemann, P. Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides. Fungal Genet. Biol. 2012, 49, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, C.; Wang, Y.; Yao, X.; Wang, F.; Wu, J.; King, G.J.; Liu, K. Disruption of a carotenoid cleavage dioxygenase 4 gene converts flower color from white to yellow in Brassica species. New Phytol. 2015, 206, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Bruno, M.; Beyer, P.; Al-Babili, S. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 2015, 572, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Soares, V.L.; Rodrigues, S.M.; de Oliveira, T.M.; de Queiroz, T.O.; Lima, L.S.; Hora-Júnior, B.T.; Gramacho, K.P.; Micheli, F.; Cascardo, J.C.; Otoni, W.C. Unraveling new genes associated with seed development and metabolism in Bixa orellana l. By expressed sequence tag (EST) analysis. Mol. Biol. Rep. 2011, 38, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Park, S.U. Molecular cloning and characterization of cDNAs encoding carotenoid cleavage dioxygenase in bitter melon (Momordica charantia). J. Plant Physiol. 2013, 170, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Hai, N.T.L.; Masuda, J.-I.; Miyajima, I.; Thien, N.Q.; Mojtahedi, N.; Hiramatsu, M.; Kim, J.-H.; Okubo, H. Involvement of carotenoid cleavage dioxygenase 4 gene in tepal color change in Lilium brownii var. colchesteri. J. Jpn. Soc. Hort. Sci. 2012, 81, 366–373. [Google Scholar] [CrossRef]
- Obrero, A.; González-Verdejo, C.I.; Die, J.V.; Gómez, P.; Del Río-Celestino, M.; Román, B.N. Carotenogenic gene expression and carotenoid accumulation in three varieties of Cucurbita pepo during fruit development. J. Agric. Food Chem. 2013, 61, 6393–6403. [Google Scholar] [CrossRef] [PubMed]
- Brandi, F.; Bar, E.; Mourgues, F.; Horváth, G.; Turcsi, E.; Giuliano, G.; Liverani, A.; Tartarini, S.; Lewinsohn, E.; Rosati, C. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 2011, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Adami, M.; De Franceschi, P.; Brandi, F.; Liverani, A.; Giovannini, D.; Rosati, C.; Dondini, L.; Tartarini, S. Identifying a carotenoid cleavage dioxygenase (ccd4) gene controlling yellow/white fruit flesh color of peach. Plant Mol. Biol. Rep. 2013, 31, 1166–1175. [Google Scholar] [CrossRef]
- Falchi, R.; Vendramin, E.; Zanon, L.; Scalabrin, S.; Cipriani, G.; Verde, I.; Vizzotto, G.; Morgante, M. Three distinct mutational mechanisms acting on a single gene underpin the origin of yellow flesh in peach. Plant J. 2013, 76, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Fukamatsu, Y.; Tamura, T.; Hihara, S.; Oda, K. Mutations in the CCD4 carotenoid cleavage dioxygenase gene of yellow-flesh peaches. Biosci. Biotechnol. Biochem. 2013, 77, 2514–1526. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, J.; Zhao, J.; Zhou, H.; Ren, F.; Wang, L.; Gu, C.; Liao, L.; Han, Y. Inactivation of a gene encoding carotenoid cleavage dioxygenase (ccd4) leads to carotenoid-based yellow coloration of fruit flesh and leaf midvein in peach. Plant Mol. Biol. Rep. 2014, 32, 246–257. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, S.; Sun, W.; Wu, M.; Hu, W.; Shen, X.; Wang, Y. Comparative analysis of carotenoid accumulation in two goji (Lycium barbarum L. and L. ruthenicum murr.) fruits. BMC Plant Biol. 2014, 14, 269. [Google Scholar] [CrossRef] [PubMed]
- Ramel, F.; Mialoundama, A.S.; Havaux, M. Nonenzymic carotenoid oxidation and photooxidative stress signalling in plants. J. Exp. Bot. 2013, 64, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Sorefan, K.; Booker, J.; Haurogne, K.; Goussot, M.; Bainbridge, K.; Foo, E.; Chatfield, S.; Ward, S.; Beveridge, C.; Rameau, C.; et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Qin, X.; Loewen, M.C. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J. Biol. Chem. 2004, 279, 46940–46945. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Roldán, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Walter, M.H.; Giavalisco, P.; Lytovchenko, A.; Kohlen, W.; Charnikhova, T.; Simkin, A.J.; Goulet, C.; Strack, D.; Bouwmeester, H.J.; et al. Slccd7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010, 61, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H.; LekKala, S.P.; Bhattacharya, C.; Mayzlish-Gati, E.; Resnick, N.; Wininger, S.; Dor, E.; Yoneyama, K.; Hershenhorn, J.; Joel, D.M.; et al. A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 2010, 61, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, H.J.; Roux, C.; López-Raez, J.A.; Becard, G. Rhizosphere communication of plants, parasitic plants and am fungi. Trends Plant Sci. 2007, 12, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Waldie, T.; McCulloch, H.; Leyser, O. Strigolactones and the control of plant development: Lessons from shoot branching. Plant J. 2014, 79, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.; Ruyter-Spira, C.; Bouwmeester, H.J. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 2011, 180, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of witchweed (Striga lutea lour.): Isolation and properties of a potent stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yoneyama, K. The strigolactone story. Annu. Rev. Phytopathol. 2010, 48, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Shirasu, K. Plants that attack plants: Molecular elucidation of plant parasitism. Curr. Opin. Plant Biol. 2012, 15, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Jamil, M.; Charnikhova, T.; Verstappen, F.; Bouwmeester, H. Carotenoid inhibitors reduce strigolactone production and Striga hermonthica infection in rice. Arch. Biochem. Biophys. 2010, 504, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. D14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, K.; Sorefan, K.; Ward, S.; Leyser, O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 2005, 44, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; van Zeijl, A.; van Bezouwen, L.; de Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011, 155, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Snowden, K.C.; Simkin, A.J.; Janssen, B.J.; Templeton, K.R.; Loucas, H.M.; Simons, J.L.; Karunairetnam, S.; Gleave, A.P.; Clark, D.G.; Klee, H.J. The decreased apical dominance1/petunia hybrida carotenoid cleavage dioxygenase8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 2005, 17, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Bullier, E.; Goussot, M.; Foucher, F.; Rameau, C.; Beveridge, C.A. The branching gene ramosus1 mediates interactions among two novel signals and auxin in pea. Plant Cell 2005, 17, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Ledger, S.E.; Janssen, B.J.; Karunairetnam, S.; Wang, T.; Snowden, K.C. Modified carotenoid cleavage dioxygenase8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytol. 2010, 188, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Charnikhova, T.; Lammers, M.; Pollina, T.; Toth, P.; Haider, I.; Pozo, M.J.; de Maagd, R.A.; Ruyter-Spira, C.; Bouwmeester, H.J.; et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012, 196, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhao, L.; Challis, R.; Leyser, O. Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum). J. Exp. Bot. 2010, 61, 3069–3078. [Google Scholar] [CrossRef] [PubMed]
- Djennane, S.; Hibrand-Saint Oyant, L.; Kawamura, K.; Lalanne, D.; Laffaire, M.; Thouroude, T.; Chalain, S.; Sakr, S.; Boumaza, R.; Foucher, F.; et al. Impacts of light and temperature on shoot branching gradient and expression of strigolactone synthesis and signalling genes in rose. Plant Cell Environ. 2013, 37, 742–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.S.; Jeon, Y.A.; Lim, S.H.; Kim, J.K.; Lee, J.Y.; Kim, Y.M.; Lee, Y.H.; Ha, S.H. Vascular-specific activity of the Arabidopsis carotenoid cleavage dioxygenase 7 gene promoter. Plant Cell Rep. 2011, 30, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhao, X.; Pan, X.; Xie, Q.; Zhu, L. The rice high-tillering DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006, 48, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Drummond, R.S.; Martinez-Sanchez, N.M.; Janssen, B.J.; Templeton, K.R.; Simons, J.L.; Quinn, B.D.; Karunairetnam, S.; Snowden, K.C. Petunia hybrida carotenoid cleavage dioxygenase7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 2009, 151, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Ruyter-Spira, C.; Bouwmeester, H. The interaction between strigolactones and other plant hormones in the regulation of plant development. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Delaux, P.M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Sejalon-Delmas, N.; Combier, J.P.; Becard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, A.; Waters, M.T.; Ghisalberti, E.L.; Dixon, K.W.; Flematti, G.R.; Smith, S.M. Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 2013, 76, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Novero, M.; Charnikhova, T.; Ferrandino, A.; Schubert, A.; Ruyter-Spira, C.; Bonfante, P.; Lovisolo, C.; Bouwmeester, H.J.; Cardinale, F. CAROTENOID CLEAVAGE DIOXYGENASE 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 2013, 64, 1967–1981. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Saika, H.; Maekawa, M.; Takamure, I.; Tsutsumi, N.; Kyozuka, J.; Nakazono, M. Rice tillering dwarf mutant dwarf3 has increased leaf longevity during darkness-induced senescence or hydrogen peroxide-induced cell death. Genes Genet. Syst. 2007, 82, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X.; Brcich, T.; Dun, E.A.; Goussot, M.; Haurogne, K.; Beveridge, C.A.; Rameau, C. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 2006, 142, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Foo, E. Auxin influences strigolactones in pea mycorrhizal symbiosis. J. Plant Physiol. 2013, 170, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Moraga, A.; Ahrazem, O.; Pérez-Clemente, R.M.; Gómez-Cadenas, A.; Yoneyama, K.; López-Raez, J.A.; Molina, R.V.; Gómez-Gómez, L. Apical dominance in saffron and the involvement of the branching enzymes CDD7 and CCD8 in the control of bud sprouting. BMC Plant Biol. 2014, 14, 171. [Google Scholar] [CrossRef] [PubMed]
- Liaaen-Jensen, S. Carotenoids in chemosystematics. In Biosynthesis and Metabolism; Britton, G., Liaaen-Jensen, S., Pfander, H., Eds.; Birkhäuser: Basel, Switzerland, 1998. [Google Scholar]
- Takaichi, S. Carotenoids in algae: Distributions, biosyntheses and functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, G.; Marner, F.J.; Brohsonn, U.; Melkonian, M. Identification of 11-cis and all-trans-retinal in the photoreceptive organelle of a flagellate green alga. FEBS Lett. 1991, 293, 49–52. [Google Scholar] [CrossRef]
- Derguini, F.; Mazur, P.; Nakanishi, K.; Starace, D.M.; Saranak, J.; Foster, K.W. All-trans-retinal is the chromophore bound to the photoreceptor of the alga Chlamydomonas reinhardtii. Photochem. Photobiol. 1991, 54, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Sineshchekov, O.A.; Govorunova, E.G.; Jung, K.H.; Zauner, S.; Maier, U.G.; Spudich, J.L. Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys. J. 2005, 89, 4310–4319. [Google Scholar] [CrossRef] [PubMed]
- Sineshchekov, O.A.; Jung, K.H.; Spudich, J.L. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2002, 99, 8689–8694. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Gessner, G.; Luff, M.; Heiland, I.; Wagner, V.; Kaminski, M.; Geimer, S.; Eitzinger, N.; Reissenweber, T.; Voytsekh, O.; et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 2006, 18, 1908–1930. [Google Scholar] [CrossRef] [PubMed]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.P.; Yuan, J.; Sun, L.; She, Z.G.; Wu, J.H.; Lan, X.J.; Zhu, X.; Lin, Y.C.; Chen, S.P. Statistical research on marine natural products based on data obtained between 1985 and 2008. Mar. Drugs 2011, 9, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Delaux, P.M.; Xie, X.; Timme, R.E.; Puech-Pages, V.; Dunand, C.; Lecompte, E.; Delwiche, C.F.; Yoneyama, K.; Becard, G.; Sejalon-Delmas, N. Origin of strigolactones in the green lineage. New Phytol. 2012, 195, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Baldermann, S.; Yoshikawa, K.; Fujita, A.; Mase, N.; Watanabe, N. Determination of volatile compounds in four commercial samples of Japanese green algae using solid phase microextraction gas chromatography mass spectrometry. Sci. World J. 2014, 2014, 289780. [Google Scholar] [CrossRef] [PubMed]
- Baldermann, S.; Yamamoto, M.; Yang, Z.; Kawahashi, T.; Kuwano, K.; Watanabe, N. C13-apocarotenoids: More than flavor compounds? In Carotenoid Cleavage Products; Winterhalter, P., Ebeler, S.E., Eds.; ACS Symposium Series: Washington, DC, USA, 2013. [Google Scholar]
- Ikawa, M.; Sasner, J.J.; Haney, J.F. Activity of cyanobacterial and algal odor compounds found in lake waters on green alga Chlorella pyrenoidosa growth. Hydrobiologia 2001, 443, 19–22. [Google Scholar] [CrossRef]
- Baldermann, S.; Mulyadi, A.N.; Yang, Z.; Murata, A.; Fleischmann, P.; Winterhalter, P.; Knight, M.; Finn, T.M.; Watanabe, N. Application of centrifugal precipitation chromatography and high-speed counter-current chromatography equipped with a spiral tubing support rotor for the isolation and partial characterization of carotenoid cleavage-like enzymes in Enteromorpha compressa (L.) nees. J. Sep. Sci. 2011, 34, 2759–2764. [Google Scholar] [PubMed]



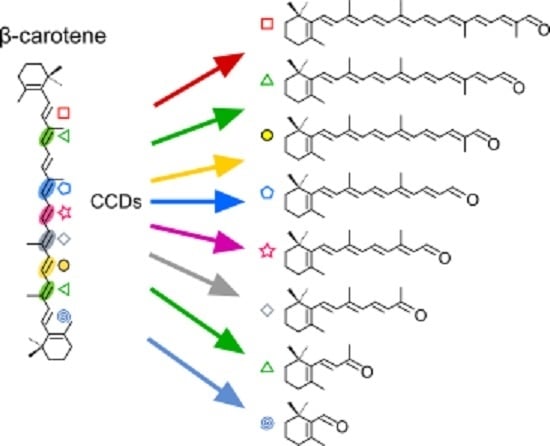
 : C5–C6;
: C5–C6;  : C7–C8;
: C7–C8;  : C9–C10 and C9′–C10′;
: C9–C10 and C9′–C10′;  : C13–C14;
: C13–C14;  : C13′–C14′;
: C13′–C14′;  : C11′–C12′.
: C11′–C12′.
 : C5–C6;
: C5–C6;  : C7–C8;
: C7–C8;  : C9–C10 and C9′–C10′;
: C9–C10 and C9′–C10′;  : C13–C14;
: C13–C14;  : C13′–C14′;
: C13′–C14′;  : C11′–C12′.
: C11′–C12′.

| Substrates | Diox1 Synechocystis sp. PCC 6803 | NosAco Nostoc sp. PCC 7120 |
|---|---|---|
β-apo-4′-carotenal | Retinal | Not detected |
β-apo-8′-carotenal | Retinal | Retinal |
β-apo-10′-carotenal | Retinal | Retinal |
β-apo-12′-carotenal | Retinal | Not detected |
(3R)-3-OH-β-apo-8′-carotenal | 3-OH-retinal | 3R-3-OH-retinal |
(3R)-3-OH-β-apo-12′-carotenal | 3-OH-retinal | 3R-3-OH-retinal |
Apo-8′-lycopenal | Acycloretinal (slow) | Acycloretinal |
Apo-10′-lycopenal | No information | Acycloretinal |
β-apo-8′-carotenol | Retinal | Retinal |
(3R)-3-OH-β-apo-8′-carotenol | 3-OH-retinal | 3-OH-retinal |
Apo-8′-lycopenol | No information | Acycloretinal |
4-oxo-β-apo-8′-carotenal | 4-oxo-retinal (slow) | Not tested |
| References | [29] | [32] |
 : Enzyme cleaves at the C15–C15′ double bond, or equivalent positions.
: Enzyme cleaves at the C15–C15′ double bond, or equivalent positions.| Assayed Substrates | Enzymes and Products | ||||||
|---|---|---|---|---|---|---|---|
| β-carotene-oxygenase Microcystis PCC 7806 | NosCCD (NSC1) Nostoc sp. PCC 7120 | MtCCO Mycobacterium tuberculosis | NSC3 Nostoc sp. PCC 7120 | NACOX1 Novosphingobium | SaCCO Sphingopyxis alaskensis | PpCCO Plesiocystis pacifica | |
β-apo-8′-carotenal | β-ionone (  ) + Apo-8,10′-apocarotene-dial ( ) + Apo-8,10′-apocarotene-dial (  ) )Other minority products (  , ,  ) ) | β-apo-13-carotenone (  ) )Retinal (  ) ) | β-apo-13-carotenone (  ) )β-apo-14′-carotenal Transretinal (  ) ) | β-apo-13-carotenone (  ) ) | Apo-12′-carotenal (  ) )Apo-10′-carotenal (  ) ) | ||
β-apo-10′-carotenal | β-ionone (  ) + Apo-10,10′-apocarotene-dial ( ) + Apo-10,10′-apocarotene-dial (  ) ) | β-apo-13-carotenone (  ) )Retinal (  ) ) | |||||
3-OH-β-apo-8′-carotenal | 3-OH-β-ionone (  ) + Apo-8,10′-apocarotene-dial ( ) + Apo-8,10′-apocarotene-dial (  ) ) | 3-OH-β-apo-13-carotenone (  ) )3-OH-retinal (  ) ) | |||||
3-OH-β-apo-10′-carotenal | 3-OH-β-ionone (  ) + Apo-10,10′-apocarotene-dial ( ) + Apo-10,10′-apocarotene-dial (  ) ) | 3-OH-β-apo-13-carotenone (  ) )3-OH-retinal (  ) ) | |||||
γ-carotene | β-ionone (  ) + Apo-8,10′-apocarotene-dial ( ) + Apo-8,10′-apocarotene-dial (  ) ) | β-apo-13-carotenone (  ) ) | |||||
β-carotene | 2 × β-cyclocitral (  , ,  ) )Crocetindial (  , ,  ) ) | Apo-10,10′-apocarotene-dial (  ) ) | β-apo-13-carotenone (  ) )Retinal (  ) )β-apo-14′-carotenal (  ) ) | No | No | No (in vitro) | No |
Zeaxanthin | 2 × hydroxi-β-cyclocitral (  , ,  ) )Crocetindial (  , ,  ) ) | 3-OH-β-ionone (  ) )Apo-10,10′-apocarotene-dial (  ) ) | 3-OH-β-apo-13-carotenone (  ) )3-OH-β-apo-14′-carotenal (  ) )3-OH-retinal (  ) )3-OH-β-apo-11-carotenal? | No | No | No (in vitro) | Apo-13′-zeaxanthinone (  ) )Apo-14′-zeaxanthinal (  ) )Apo-12′-zeaxanthinal (  ) ) |
Lycopene | No | No | poorly | Yes (in vivo) Unknown product | |||
Lutein | 3-OH-β-apo-13-carotenone (  ) )3-OH-β-apo-14′-carotenal (  ) )3-OH-retinal (  ) )3-OH-α-apo-15′-carotenal(  ) ) | No | |||||
Echinenone | No | 4-oxo-β-ionone (  ) )Apo-10,10′-apocarotene-dial (  ) )β-ionone (  ) ) | |||||
Canthaxantin | 2 x 4-oxo-β-ionone (  ) )Apo-10,10′-apocarotene-dial (  ) ) | Apo-13′-canta-xanthinone (  ) )Apo-14′-cantaxanthinal (  ) )Apo-12′-cantaxanthinal (  ) ) | |||||
Astaxanthin | 3-OH-β-ionone (  ) )3-OH, 4-oxo-β-inone (  ) )Apo-10,10′-apocarotene-dial (  ) ) | Apo-13′-astaxanthinone (  ) )Apo-14′-astaxanthinal (  ) ) | |||||
| References | [19] | [30,33] | [36] | [31] | [37] | [38] | [38] |
 : C7–C8;
: C7–C8;  : C9–C10;
: C9–C10;  : C13–C14;
: C13–C14;  : C15–C15′;
: C15–C15′;  : C13′–C14′;
: C13′–C14′;  : C11′-C12′;
: C11′-C12′;  : C9′–C10′;
: C9′–C10′;  : C7′–C8′. When an enzyme cleaves at different positions, the predominant product is indicated in bold. Lack of bold means no product preference or information not available.
: C7′–C8′. When an enzyme cleaves at different positions, the predominant product is indicated in bold. Lack of bold means no product preference or information not available.| Substrates | Oxygenase | Products |
|---|---|---|
β-apo-4′-carotenal | NACOX1 | β-apo-13-carotenone (  ) ) |
3,3′-dihydroxy-isorenieratene | MtCCO | 3-OH-β-apo-13-carotenone (  ) )3-OH-β-apo-15′-carotenal (  ) )3-OH-β-apo-14′-carotenal (  ) ) |
Echinenone | NosCCD | Apo-10,10′-apocarotenoid (  ) )2 × C13 (  ) ) |
Nostoxanthin | PpCCO | Apo-14′-nostoxanthinal (  ) )Apo-12′-nostoxanthinal (  ) ) |
Hydroxylycopene | PpCCO | Unknown |
Dihydroxylycopene | PpCCO | Unknown |
Torulene | NSC3 | Transretinal (  ) ) |
4,4′-diapotorulene | NSC3 | Apo-14′-diapotorulenal (  ) ) |
4,4′-diapotorulene-4-al | NSC3 | Apo-10′-diapotorulenal (  ) ) |
4,4′-diaponeurosporene | NSC3 | Apo-14′-diaponeurosporenal (  ) ) |
4,4′-diaponeurosporen-4′-al | NSC3 | Apo-14′-diaponeurosporenal (  ) )Apo-10′-diaponeurosporenal (  ) ) |
4,4′-diaponeurosporen-4′-oic acid | NSC3 | Apo-10′-diaponeurosporenal (  ) ) |
Myxol | NosCCD | 3-OH-β-ionone (  ) )Apo-8′-10-apocarotenedial (  ) ) |
Myxol-2′-fucoside | NosCCD | 3-OH-β-ionone (  ) )Apo-8′-10-apocarotenedial (  ) )acetate adduct C10 |
4-ketomyxol-2′-fucoside | NosCCD | 3-OH-4-oxo-β-ionone (  ) )Apo-8′-10-apocarotenedial (  ) ) |
4-hydroxymyxol-2′-fucoside | NosCCD | 3, 4-OH-β-ionone (  ) )Apo-8′-10-apocarotenedial (  ) ) |
 : C9=C10;
: C9=C10;  : C13=C14;
: C13=C14;  : C15=C15′;
: C15=C15′;  : C13′=C14′;
: C13′=C14′;  : C11′=C12′;
: C11′=C12′;  : C9′=C10′;
: C9′=C10′;  : C7′=C8′.
: C7′=C8′.| Species | Protein | Assay a | Parent Carotenoid | Cleavage Position | Product Detected | References |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | CCD4 | in planta | β-carotene, Violaxanthin | n.d. | n.d. | [107] |
| in planta | Phytofluene, ζ-carotene | n.d. | n.d. | [135] | ||
| in planta (leaf) | Epoxy-β-xanthophylls | C9–C10 or C9′–C10′ | C13-glycosids | [134] | ||
| in planta (root) | β-carotene | n.d. | long-chain free apocarotenals (C15 to C30) | [134] | ||
| in vitro | Apo-β-caroten-8′-al | C9–C10 | β-ionone | [96] | ||
| Brassica napus | CCD4_C3 | in planta | α-carotene, δ-carotene | C9–C10 | α-ionone | [152] |
| Chrysantemum morifolium | CCD4a | in vivo and in vitro | β-carotene | C9–C10 (C9′–C10′) | β-ionone | [96] |
| Citrus clementina | CCD4b1 | in vitro | β-carotene, β-cryptoxanthin, Zeaxanthin, Lutein, α-carotene | C7–C8 or C7′–C8′ | 3-OH-apo-β-8-carotenal, apo-β-8-carotenal (C30); β-cyclocitral and 3-OH-β-cyclocitral (C10) | [133] |
| Citrus unshiu | CCD4 | in vitro and in vivo | β-cryptoxanthin, Zeaxanthin | C7–C8 or C7′–C8′ | 3-OH-apo-β-8-carotenal | [131] |
| Crocus sativus | CCD4a/b | in vivo | β-carotene, Zeaxanthin | C9–C10 (C9′–C10′) | β-ionone, β-OH-ionone | [89,147] |
| CCD4c | in vivo | β-carotene | C9–C10 or C9′–C10′ | β-ionone, β-cyclocitral | [130] | |
| in planta | C7–C8 or C7′–C8′ | |||||
| in planta | Lutein | C9–C10 | Megastigma-4,6,8-triene (derived from 3-OH-α-ionone) | [130] | ||
| Malus domestica | CCD4 | in vivo | β-carotene | C9–C10 (C9′–C10′) | β-ionone | [96] |
| Osmanthus fragans | CCD4 | in vivo | β-carotene | C9–C10 (C9′–C10′) | β-ionone | [96] |
| Rosa damascena | CCD4 | in vivo | β-carotene | C9–C10 (C9′–C10′) | β-ionone | [96] |
| in vitro | apo-β-8-carotenal | C9–C10 | β-ionone | [96] | ||
| Solanum tuberosum | CCD4 | in planta | Violaxanthin | n.d. | n.d. | [150] |
| in vitro and in vivo | β-carotene | C9–C10 or C9′–C10′ | Apo-β-caroten-10′-al; β-ionone | [153] | ||
| in vitro | α-carotene, Lutein, Zeaxanthin, β-cryptoxanthin | C9–C10 or C9′–C10′ | 3-OH-β-apo-10′-carotenal, β-apo-10′-carotenal3-OH-ε-apo-10´-carotenal | [153] | ||
| Vitis vinifera | CCD4a/b | in vivo | ε-carotene, | C9–C10 (C9′–C10′) | α-ionone, | [98] |
| CCD4a/b | in vivo | Neurosporene | C9′–C10′ | Geranylacetone | [98] | |
| CCD4a/b | in vivo | Lycopene | C5–C6 (C5′–C6′) | 6-methyl-5-hepten-2-one | [98] | |
| CCD4b | in vivo | ζ-carotene | C9–C10 (C9′–C10′) | α-ionone, Geranylacetone | [98] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahrazem, O.; Gómez-Gómez, L.; Rodrigo, M.J.; Avalos, J.; Limón, M.C. Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions. Int. J. Mol. Sci. 2016, 17, 1781. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111781
Ahrazem O, Gómez-Gómez L, Rodrigo MJ, Avalos J, Limón MC. Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions. International Journal of Molecular Sciences. 2016; 17(11):1781. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111781
Chicago/Turabian StyleAhrazem, Oussama, Lourdes Gómez-Gómez, María J. Rodrigo, Javier Avalos, and María Carmen Limón. 2016. "Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions" International Journal of Molecular Sciences 17, no. 11: 1781. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111781