Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery
Abstract
:1. Introduction
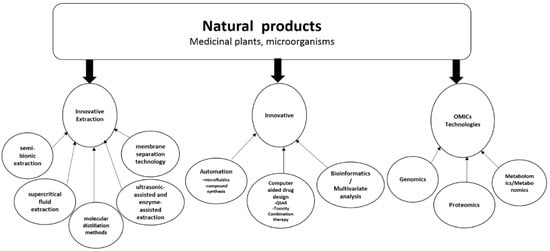
2. Multidisciplinary Approach to Natural Products Drug Discovery Using Innovative Technologies
3. Natural Products Drug Discovery Research and Development and Omics (Genomics Proteomics and Metabolomics/Metabonomics)
3.1. Genomics in Plant-Based Natural Products Identification and Biomarker Identification
3.2. Proteomics in Natural Product Validation and Biomarker Identification
Methods for Target Identification of Label-Free Natural Products
3.3. Metabolomics and Metabonomics Approach to Natural Products Drug Discovery
3.4. Big Data in Drug Development for Natural Product Drug Development and Precision Medicine
4. Automating Natural Product Drug Discovery
5. Computer-Aided Drug Design from Natural Products
6. Natural Products and Precision Medicine
7. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Weng, J.K.; Philippe, R.N.; Noel, J.P. The rise of chemodiversity in plants. Science 2012, 336, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Lietava, J. Medicinal plants in a Middle Paleolithic grave Shanidar IV? J. Ethnopharmacol. 1992, 35, 263–266. [Google Scholar] [CrossRef]
- Ernst, M.; Grace, O.M.; Saslis-Lagoudakis, C.H.; Nilsson, N.; Simonsen, H.T.; Ronsted, N. Global medicinal uses of Euphorbia L. (Euphorbiaceae). J. Ethnopharmacol. 2015, 176, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Gozubuyuk, G.S.; Aktas, E.; Yigit, N. An ancient plant Lawsonia inermis (henna): Determination of in vitro antifungal activity against dermatophytes species. J. Mycol. Med. 2014, 24, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Hotwani, K.; Baliga, S.; Sharma, K. Phytodentistry: Use of medicinal plants. J. Complement. Integr. Med. 2014, 11, 233–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Lawrence, A.J.; Liang, J.H. Traditional Chinese medicine for treatment of alcoholism: From ancient to modern. Am. J. Chin. Med. 2011, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mannangatti, P.; Naidu, K.N. Indian herbs for the treatment of neurodegenerative disease. Adv. Neurobiol. 2016, 12, 323–336. [Google Scholar] [PubMed]
- McGovern, P.E.; Mirzoian, A.; Hall, G.R. Ancient Egyptian herbal wines. Proc. Natl. Acad. Sci. USA 2009, 106, 7361–7366. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L. Natural products in drug discovery. Drug Discov. Today 2008, 13, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Clark, R.L.; Mackay, S.P.; Johnston, B.F. Current strategies for drug discovery through natural products. Expert Opin. Drug Discov. 2010, 5, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.; Kim, Y.; Kwon, H.J. Advances in identification and validation of protein targets of natural products without chemical modification. Nat. Prod. Rep. 2016, 33, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Tansaz, M.; Tajadini, H. Comparison of leiomyoma of modern medicine and traditional Persian medicine. J. Evid.-Based Complement. Altern. Med. 2016, 21, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Bauer, R.; Hendry, B.M.; Fan, T.P.; Zhao, Z.; Duez, P.; Simmonds, M.S.; Witt, C.M.; Lu, A.; Robinson, N.; et al. The quest for modernisation of traditional Chinese medicine. BMC Complement. Altern. Med. 2013, 13, 132. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The traditional medicine and modern medicine from natural products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Banjari, I.; Misir, A.; Savikin, K.; Jokic, S.; Molnar, M.; De Zoysa, H.K.S.; Waisundara, V.Y. Antidiabetic effects of Aronia melanocarpa and its other therapeutic properties. Front. Nutr. 2017, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Yatoo, M.I.; Dimri, U.; Gopalakrishnan, A.; Karthik, K.; Gopi, M.; Khandia, R.; Saminathan, M.; Saxena, A.; Alagawany, M.; Farag, M.R.; et al. Beneficial health applications and medicinal values of pedicularis plants: A review. Biomed. Pharmacother. 2017, 95, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Thomford, N.E.; Awortwe, C.; Dzobo, K.; Adu, F.; Chopera, D.; Wonkam, A.; Skelton, M.; Blackhurst, D.; Dandara, C. Inhibition of cyp2b6 by medicinal plant extracts: Implication for use of efavirenz and nevirapine-based highly active anti-retroviral therapy (HAART) in resource-limited settings. Molecules 2016, 21, 211. [Google Scholar] [CrossRef] [PubMed]
- Thomford, N.E.; Dzobo, K.; Chopera, D.; Wonkam, A.; Maroyi, A.; Blackhurst, D.; Dandara, C. In vitro reversible and time-dependent cyp450 inhibition profiles of medicinal herbal plant extracts Newbouldia laevis and Cassia abbreviata: Implications for herb-drug interactions. Molecules 2016, 21, 891. [Google Scholar] [CrossRef] [PubMed]
- Thomford, N.E.; Dzobo, K.; Chopera, D.; Wonkam, A.; Skelton, M.; Blackhurst, D.; Chirikure, S.; Dandara, C. Pharmacogenomics implications of using herbal medicinal plants on African populations in health transition. Pharmaceuticals 2015, 8, 637–663. [Google Scholar] [CrossRef] [PubMed]
- Thomford, N.E.; Mkhize, B.; Dzobo, K.; Mpye, K.; Rowe, A.; Parker, M.I.; Wonkam, A.; Skelton, M.; September, A.V.; Dandara, C. African lettuce (Launaea taraxacifolia) displays possible anticancer effects and herb-drug interaction potential by CYP1A2, CYP2C9, and CYP2C19 inhibition. Omics 2016, 20, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Fattahi, A.; Raffel, N.; Hoffmann, I.; Beckmann, M.W.; Dittrich, R.; Schrauder, M. Antioxidant effect of aqueous extract of four plants with therapeutic potential on gynecological diseases; semen persicae, Leonurus cardiaca, Hedyotis diffusa, and Curcuma zedoaria. Eur. J. Med. Res. 2017, 22, 50. [Google Scholar] [CrossRef] [PubMed]
- Ruhsam, M.; Hollingsworth, P.M. Authentication of eleutherococcus and rhodiola herbal supplement products in the United Kingdom. J. Pharm. Biomed. Anal. 2017, 149, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.C.; Taylor, H.L.; Wall, M.E.; Coggon, P.; McPhail, A.T. Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 1971, 93, 2325–2327. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.T. Natural products and Pharma 2011: Strategic changes spur new opportunities. Nat. Prod. Rep. 2011, 28, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Li, F.S.; Weng, J.K. Demystifying traditional herbal medicine with modern approach. Nat. Plants 2017, 3, 17109. [Google Scholar] [CrossRef] [PubMed]
- Kiyohara, H.; Matsumoto, T.; Yamada, H. Combination effects of herbs in a multi-herbal formula: Expression of juzen-taiho-to’s immuno-modulatory activity on the intestinal immune system. Evid.-Based Complement. Altern. Med. 2004, 1, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Leonti, M.; Verpoorte, R. Traditional Mediterranean and European herbal medicines. J. Ethnopharmacol. 2017, 199, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, V.; Hekim, N. Birth of industry 5.0: Making sense of big data with artificial intelligence, “the internet of things” and next-generation technology policy. Omics 2018, 22, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, V. Omics 2.0: An accelerator for global science, systems medicine and responsible innovation. Omics 2015, 19, 579–580. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.E.; Wani, M.C.; Brown, D.M.; Fullas, F.; Olwald, J.B.; Josephson, F.F.; Thornton, N.M.; Pezzuto, J.M.; Beecher, C.W.; Farnsworth, N.R.; et al. Effect of tannins on screening of plant extracts for enzyme inhibitory activity and techniques for their removal. Phytomedicine 1996, 3, 281–285. [Google Scholar] [CrossRef]
- Eldridge, G.R.; Vervoort, H.C.; Lee, C.M.; Cremin, P.A.; Williams, C.T.; Hart, S.M.; Goering, M.G.; O’Neil-Johnson, M.; Zeng, L. High-throughput method for the production and analysis of large natural product libraries for drug discovery. Anal. Chem. 2002, 74, 3963–3971. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liang, J. Counter-current chromatography for high throughput analysis of natural products. Comb. Chem. High Throughput Screen. 2010, 13, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Richards, B.; Bhoite, L.; Cimbora, D.; Harper, M.K.; Ireland, C.M. Marine natural product libraries for high-throughput screening and rapid drug discovery. J. Nat. Prod. 2008, 71, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Koehn, F.E. High impact technologies for natural products screening. In Natural Compounds as Drugs Volume I; Birkhäuser: Basel, Switzerland, 2008; Volume 65, pp. 175–210. [Google Scholar]
- Wong, W.R.; Oliver, A.G.; Linington, R.G. Development of antibiotic activity profile screening for the classification and discovery of natural product antibiotics. Chem. Biol. 2012, 19, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Yin, Y.; Yan, X.; Wang, Y. Semi-bionic extraction of effective ingredient from fishbone by high intensity pulsed electric fields. J. Food Process Eng. 2017, 40, e12392. [Google Scholar] [CrossRef]
- Yoshioka, T.; Nagatomi, Y.; Harayama, K.; Bamba, T. Development of an analytical method for polycyclic aromatic hydrocarbons in coffee beverages and dark beer using novel high-sensitivity technique of supercritical fluid chromatography/mass spectrometry. J. Biosci. Bioeng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hofstetter, R.; Fassauer, G.M.; Link, A. Supercritical fluid extraction (SFE) of ketamine metabolites from dried urine and on-line quantification by supercritical fluid chromatography and single mass detection (on-line SFE–SFC–MS). J. Chromatogr. B 2018, 1076, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.; Piris, A.J.; Ruiz-Rodriguez, A.; Prodanov, M.; Soler-Rivas, C. Extraction of bioactive compounds against cardiovascular diseases from Lentinula edodes using a sequential extraction method. Biotechnol. Prog. 2018. [Google Scholar] [CrossRef] [PubMed]
- Joana Gil-Chávez, G.; Villa, J.A.; Fernando Ayala-Zavala, J.; Basilio Heredia, J.; Sepulveda, D.; Yahia, E.M.; González-Aguilar, G.A. Technologies for extraction and production of bioactive compounds to be used as nutraceuticals and food ingredients: An overview. Compr. Rev. Food Sci. Food Saf. 2013, 12, 5–23. [Google Scholar] [CrossRef]
- De Morais, S.R.; Oliveira, T.L.; de Oliveira, L.P.; Tresvenzol, L.M.; da Conceicao, E.C.; Rezende, M.H.; Fiuza, T.S.; Costa, E.A.; Ferri, P.H.; de Paula, J.R. Essential oil composition, antimicrobial and pharmacological activities of Lippia sidoides cham. (verbenaceae) from Sao Goncalo do Abaete, Minas Gerais, Brazil. Pharmacogn. Mag. 2016, 12, 262–270. [Google Scholar] [PubMed]
- Gan, Z.; Liang, Z.; Chen, X.; Wen, X.; Wang, Y.; Li, M.; Ni, Y. Separation and preparation of 6-gingerol from molecular distillation residue of Yunnan ginger rhizomes by high-speed counter-current chromatography and the antioxidant activity of ginger oils in vitro. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1011, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mei, J.; Xie, Y.; Li, M.; Liu, D.; He, C. Application of membrane separation technology in extraction process of Chuanxiong Chatiao granules. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi = China J. Chin. Mater. Med. 2012, 37, 934–936. [Google Scholar]
- Williams, S.; Oatley, D.; Abdrahman, A.; Butt, T.; Nash, R. Membrane technology for the improved separation of bioactive compounds. Procedia Eng. 2012, 44, 2112–2114. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, Y.; Ding, M.; Li, J.; Hao, J.; He, J.; Wang, H.; Gao, X.M.; Chang, Y.X. Simultaneous determination and qualitative analysis of six types of components in Naoxintong capsule by miniaturized matrix solid-phase dispersion extraction coupled with ultra high-performance liquid chromatography with photodiode array detection and quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 2018. [Google Scholar] [CrossRef]
- Zhang, L.; Ge, Y.; Li, J.; Hao, J.; Wang, H.; He, J.; Gao, X.M.; Chang, Y.X. Simultaneous determination of columbianetin-beta-d-glucopyranoside and columbianetin in a biological sample by high-performance liquid chromatography with fluorescence detection and identification of other columbianetin-beta-d-glucopyranoside metabolites by ultra high-performance liquid chromatography coupled with quadrupole-time of flight mass spectrometry. J. Pharm. Biomed. Anal. 2018, 153, 221–231. [Google Scholar] [PubMed]
- Manglik, A.; Kruse, A.C.; Kobilka, T.S.; Thian, F.S.; Mathiesen, J.M.; Sunahara, R.K.; Pardo, L.; Weis, W.I.; Kobilka, B.K.; Granier, S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature 2012, 485, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Edwards, G. Forces of habit: Drugs and the making of the modern world. Addiction 2002, 97, 608–609. [Google Scholar] [CrossRef]
- Zhao, L.; Li, C.; Zhang, Y.; Wen, Q.; Ren, D. Phytochemical and biological activities of an anticancer plant medicine: Brucea javanica. Anti-Cancer Agents Med. Chem. 2014, 14, 440–458. [Google Scholar] [CrossRef]
- Tu, Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y. Artemisinin—A gift from traditional Chinese medicine to the world (Nobel lecture). Angew. Chem. Int. Ed. Engl. 2016, 55, 10210–10226. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Casteels, T.; Frogne, T.; Ingvorsen, C.; Honoré, C.; Courtney, M.; Huber, K.V.M.; Schmitner, N.; Kimmel, R.A.; Romanov, R.A.; et al. Artemisinins target GABAA receptor signaling and impair α cell identity. Cell 2017, 168, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Singh, N.P. Oral artemisinin prevents and delays the development of 7,12-dimethylbenz[a]anthracene (dmba)-induced breast cancer in the rat. Cancer Lett. 2006, 231, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.C.; Singh, N.P.; Sasaki, T. Development of artemisinin compounds for cancer treatment. Investig. New Drugs 2013, 31, 230–246. [Google Scholar] [CrossRef] [PubMed]
- Return to Rio: Second chance for the planet. Nature 2012, 486, 19.
- Barbault, R. 2010: A new beginning for biodiversity? C.R. Biol. 2011, 334, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Salazar, R.; Cabrera, J.A. Intellectual property rights in Costa Rica in the light of the biodiversity convention. J. Ethnopharmacol. 1996, 51, 177–193. [Google Scholar] [CrossRef]
- Samper, C. Taxonomy and environmental policy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Seidl, P.R. Pharmaceuticals from natural products: Current trends. Anais da Academia Brasileira de Ciencias 2002, 74, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, J.; Gilbert, N. Earth summit: Rio report card. Nature 2012, 486, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.C.; Wei, C.H.; Hu, Z.Q.; Srivastava, K.; Ko, J.; Xi, S.T.; Mu, D.Z.; Du, J.B.; Li, G.H.; Wallenstein, S.; et al. Efficacy and tolerability of anti-asthma herbal medicine intervention in adult patients with moderate-severe allergic asthma. J. Allergy Clin. Immunol. 2005, 116, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Sampson, H.A.; Charles, W.; Emala, S.; Li, X.-M. The anti-asthma herbal medicine ashmi acutely inhibits airway smooth muscle contraction via prostaglandin e2 activation of ep2/ep4 receptors. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2013, 305, L1002–L1010. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Liang, B.; Srivastava, K.; Zeng, J.; Zhan, J.; Brown, L.; Sampson, H.; Goldfarb, J.; Emala, C.; Li, X.M. The Sophora flavescens flavonoid compound trifolirhizin inhibits acetylcholine induced airway smooth muscle contraction. Phytochemistry 2013, 95, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Shaw, D.; Simmonds, M.S.; Leon, C.J.; Xu, Q.; Lu, A.; Sutherland, I.; Ignatova, S.; Zhu, Y.P.; Verpoorte, R.; et al. Good practice in reviewing and publishing studies on herbal medicine, with special emphasis on traditional Chinese medicine and Chinese materia medica. J. Ethnopharmacol. 2012, 140, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Skroza, D.; Generalić Mekinić, I.; Svilović, S.; Šimat, V.; Katalinić, V. Investigation of the potential synergistic effect of resveratrol with other phenolic compounds: A case of binary phenolic mixtures. J. Food Compos. Anal. 2015, 38, 13–18. [Google Scholar] [CrossRef]
- Chusri, S.; Siriyong, T.; Na-Phatthalung, P.; Voravuthikunchai, S.P. Synergistic effects of ethnomedicinal plants of Apocynaceae family and antibiotics against clinical isolates of Acinetobacter baumannii. Asian Pac. J. Trop. Med. 2014, 7, 456–461. [Google Scholar] [CrossRef]
- Sharma, G.; Tyagi, A.K.; Singh, R.P.; Chan, D.C.; Agarwal, R. Synergistic anti-cancer effects of grape seed extract and conventional cytotoxic agent doxorubicin against human breast carcinoma cells. Breast Cancer Res. Treat. 2004, 85, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Moore, B.S.; Yoon, Y.J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat. Chem. Biol. 2015, 11, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Akbulut, Y.; Gaunt, H.J.; Muraki, K.; Ludlow, M.J.; Amer, M.S.; Bruns, A.; Vasudev, N.S.; Radtke, L.; Willot, M.; Hahn, S.; et al. (-)-Englerin A is a potent and selective activator of TRPC4 and TRPC5 calcium channels. Angew. Chem. Int. Ed. Engl. 2015, 54, 3787–3791. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, M.J.; Gaunt, H.J.; Rubaiy, H.N.; Musialowski, K.E.; Blythe, N.M.; Vasudev, N.S.; Muraki, K.; Beech, D.J. (-)-Englerin A-evoked cytotoxicity is mediated by Na+ influx and counteracted by Na+/K+-atpase. J. Biol. Chem. 2017, 292, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Muraki, K.; Ohnishi, K.; Takezawa, A.; Suzuki, H.; Hatano, N.; Muraki, Y.; Hamzah, N.; Foster, R.; Waldmann, H.; Nussbaumer, P.; et al. Na(+) entry through heteromeric TRPC4/C1 channels mediates (-)Englerin A-induced cytotoxicity in synovial sarcoma cells. Sci. Rep. 2017, 7, 16988. [Google Scholar] [CrossRef] [PubMed]
- Buriani, A.; Garcia-Bermejo, M.L.; Bosisio, E.; Xu, Q.; Li, H.; Dong, X.; Simmonds, M.S.; Carrara, M.; Tejedor, N.; Lucio-Cazana, J.; et al. Omic techniques in systems biology approaches to traditional Chinese medicine research: Present and future. J. Ethnopharmacol. 2012, 140, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.H.; Upadhyay, P.; Das, S.; Prasad Sharma, M. Authentication of medicinal plants by DNA markers. Plant Gene 2015, 4, 83–99. [Google Scholar] [CrossRef]
- Ghorbani, A.; Saeedi, Y.; de Boer, H.J. Unidentifiable by morphology: DNA barcoding of plant material in local markets in Iran. PLoS ONE 2017, 12, e0175722. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.A.; Newmaster, S.G. Molecular taxonomic tools provide more accurate estimates of species richness at less cost than traditional morphology-based taxonomic practices in a vegetation survey. Biodivers. Conserv. 2014, 23, 1411–1424. [Google Scholar] [CrossRef]
- Cao, M.; Wang, J.; Yao, L.; Xie, S.; Du, J.; Zhao, X. Authentication of animal signatures in traditional Chinese medicine of Lingyang Qingfei Wan using routine molecular diagnostic assays. Mol. Biol. Rep. 2014, 41, 2485–2491. [Google Scholar] [CrossRef] [PubMed]
- Newmaster, S.G.; Grguric, M.; Shanmughanandhan, D.; Ramalingam, S.; Ragupathy, S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xiang, L.; Shi, L.; Li, G.; Yao, H.; Han, J.; Lin, Y.; Song, J.; Chen, S. Identification of crude drugs in the Japanese pharmacopoeia using a DNA barcoding system. Sci. Rep. 2017, 7, 42325. [Google Scholar] [CrossRef] [PubMed]
- Pulice, G.; Pelaz, S.; Matías-Hernández, L. Molecular farming in Artemisia annua, a promising approach to improve anti-malarial drug production. Front. Plant Sci. 2016, 7, 329. [Google Scholar] [CrossRef] [PubMed]
- Gantait, S.; Debnath, S.; Nasim Ali, M. Genomic profile of the plants with pharmaceutical value. 3 Biotech 2014, 4, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Wu, X.; Wang, X.; Su, J.; Zeng, H.; Zhao, J.; Lin, S.; Liu, R.; Li, H.; Li, X.; et al. The gene expression profiles in response to 102 traditional Chinese medicine (TCM) components: A general template for research on TCMs. Sci. Rep. 2017, 7, 352. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Lo, H.-L.; Tang, W.-C.; Hsiao, H.H.-Y.; Yang, P.-M. A gene expression signature-based approach reveals the mechanisms of action of the Chinese herbal medicine Berberine. Sci. Rep. 2014, 4, 6394. [Google Scholar] [CrossRef] [PubMed]
- Kiyama, R. DNA microarray-based screening and characterization of traditional Chinese medicine. Microarrays 2017, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.K.; Liu, Y.; Lay, F.D.; Liang, G.; Berman, B.P.; Jones, P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012, 22, 2497–2506. [Google Scholar] [CrossRef] [PubMed]
- Nordlund, J.; Backlin, C.L.; Wahlberg, P.; Busche, S.; Berglund, E.C.; Eloranta, M.L.; Flaegstad, T.; Forestier, E.; Frost, B.M.; Harila-Saari, A.; et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013, 14, r105. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, Y.; Xing, X.; Liu, J.; Zhang, Y. Genome-wide analysis of DNA methylation in bovine placentas. BMC Genom. 2014, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Zykovich, A.; Hubbard, A.; Flynn, J.M.; Tarnopolsky, M.; Fraga, M.F.; Kerksick, C.; Ogborn, D.; MacNeil, L.; Mooney, S.D.; Melov, S. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell 2014, 13, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, S.; Carreira, S.; Bailey, D.; Abaitua, F.; O’Hare, P. Phosphorylation and SCF-mediated degradation regulate CREB-H transcription of metabolic targets. Mol. Biol. Cell 2015, 26, 2939–2954. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Vachhani, P.; Cortes, J.E. Treatment of relapsed/refractory acute myeloid leukemia. Curr. Treat. Options Oncol. 2017, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [PubMed]
- Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; Morozova, O.; et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Mwapagha, L.M.; Tiffin, N.; Parker, M.I. Delineation of the HPV11e6 and HPV18e6 pathways in initiating cellular transformation. Front. Oncol. 2017, 7, 258. [Google Scholar] [CrossRef] [PubMed]
- Vogelsang, M.; Wang, Y.; Veber, N.; Mwapagha, L.M.; Parker, M.I. The cumulative effects of polymorphisms in the DNA mismatch repair genes and tobacco smoking in oesophageal cancer risk. PLoS ONE 2012, 7, e36962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. Genehancer: Genome-wide integration of enhancers and target genes in genecards. Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Long, J.; Zeng, C.; Michailidou, K.; Ghoussaini, M.; Bolla, M.K.; Wang, Q.; Milne, R.L.; Shu, X.O.; Cai, Q.; et al. Fine-scale mapping of the 4q24 locus identifies two independent loci associated with breast cancer risk. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Ombrello, M.J.; Sikora, K.A.; Kastner, D.L. Genetics, genomics, and their relevance to pathology and therapy. Best Pract. Res. Clin. Rheumatol. 2014, 28, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Loomis, E.; Curry, E. DNA methylation-based chromatin compartments and CHIP-seq profiles reveal transcriptional drivers of prostate carcinogenesis. Genome Med. 2017, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Hsu, L.I.; Chiu, A.W.; Pu, Y.S.; Wang, S.H.; Liao, Y.T.; Wu, M.M.; Wang, Y.H.; Chang, C.H.; Lee, T.C.; et al. Comparison of genome-wide DNA methylation in urothelial carcinomas of patients with and without arsenic exposure. Environ. Res. 2014, 128, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mehta, G.; Jalan, R.; Mookerjee, R.P. Cracking the encode: From transcription to therapeutics. Hepatology 2013, 57, 2532–2535. [Google Scholar] [CrossRef] [PubMed]
- Tragante, V.; Moore, J.H.; Asselbergs, F.W. The encode project and perspectives on pathways. Genet. Epidemiol. 2014, 38, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Bumpus, S.B.; Evans, B.S.; Thomas, P.M.; Ntai, I.; Kelleher, N.L. A proteomics approach to discovery of natural products and their biosynthetic pathways. Nat. Biotechnol. 2009, 27, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Esteso, M.J.; Martínez-Márquez, A.; Sellés-Marchart, S.; Morante-Carriel, J.A.; Bru-Martínez, R. The role of proteomics in progressing insights into plant secondary metabolism. Front. Plant Sci. 2015, 6, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lum, J.H.; Fung, K.L.; Cheung, P.Y.; Wong, M.S.; Lee, C.H.; Kwok, F.S.; Leung, M.C.; Hui, P.K.; Lo, S.C. Proteome of oriental ginseng Panax ginseng C.A. Meyer and the potential to use it as an identification tool. Proteomics 2002, 2, 1123–1130. [Google Scholar] [CrossRef]
- Kim, S.W.; Lee, S.H.; Min, C.W.; Jo, I.H.; Bang, K.H.; Hyun, D.-Y.; Agrawal, G.K.; Rakwal, R.; Zargar, S.M.; Gupta, R.; et al. Ginseng (Panax sp.) proteomics: An update. Appl. Biol. Chem. 2017, 60, 311–320. [Google Scholar] [CrossRef]
- Li, Z.H.; Alex, D.; Siu, S.O.; Chu, I.K.; Renn, J.; Winkler, C.; Lou, S.; Tsui, S.K.; Zhao, H.Y.; Yan, W.R.; et al. Combined in vivo imaging and omics approaches reveal metabolism of icaritin and its glycosides in zebrafish larvae. Mol. BioSyst. 2011, 7, 2128–2138. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.W.; Zhang, Z.J.; Li, S.; Lei, B.; Yuan, S.; Cui, G.Z.; Man Hoi, P.; Chan, K.; Lee, S.M.Y. From omics to drug metabolism and high content screen of natural product in zebrafish: A new model for discovery of neuroactive compound. Evid.-Based Complement. Altern. Med. 2012, 2012, 605303. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.; Wang, X.; Xu, N.; Zhang, H.; Xu, H. Application of proteomics to determine the mechanism of action of traditional Chinese medicine remedies. J. Ethnopharmacol. 2014, 155, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Chen, Z. Challenges and recent advances in affinity purification of tag-free proteins. Biotechnol. Lett. 2014, 36, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Novick, D.; Rubinstein, M. Ligand affinity chromatography, an indispensable method for the purification of soluble cytokine receptors and binding proteins. Methods Mol. Biol. 2012, 820, 195–214. [Google Scholar] [PubMed]
- Pfaunmiller, E.L.; Paulemond, M.L.; Dupper, C.M.; Hage, D.S. Affinity monolith chromatography: A review of principles and recent analytical applications. Anal. Bioanal. Chem. 2013, 405, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Rix, U.; Gridling, M.; Superti-Furga, G. Compound immobilization and drug-affinity chromatography. Methods Mol. Biol. 2012, 803, 25–38. [Google Scholar] [PubMed]
- Wang, H.Z.; Chu, Z.Z.; Chen, C.C.; Cao, A.C.; Tong, X.; Ouyang, C.B.; Yuan, Q.H.; Wang, M.N.; Wu, Z.K.; Wang, H.H.; et al. Recombinant passenger proteins can be conveniently purified by one-step affinity chromatography. PLoS ONE 2015, 10, e0143598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, T.; Zhang, H.; Han, B.; Wang, L.; Kang, J. Profiling of drug binding proteins by monolithic affinity chromatography in combination with liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2014, 1359, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Schenone, M.; Dancik, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef] [PubMed]
- McFedries, A.; Schwaid, A.; Saghatelian, A. Methods for the elucidation of protein-small molecule interactions. Chem. Biol. 2013, 20, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Rix, U.; Superti-Furga, G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009, 5, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, J.W. Target identification for biologically active small molecules using chemical biology approaches. Arch. Pharm. Res. 2016, 39, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Franken, H.; Mathieson, T.; Childs, D.; Sweetman, G.M.; Werner, T.; Togel, I.; Doce, C.; Gade, S.; Bantscheff, M.; Drewes, G.; et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015, 10, 1567–1593. [Google Scholar] [CrossRef] [PubMed]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Martinez Molina, D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wang, J.; Wu, R.P.; Gomez, F.; Loo, J.A.; et al. Target identification using drug affinity responsive target stability (darts). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef] [PubMed]
- Schirle, M.; Bantscheff, M.; Kuster, B. Mass spectrometry-based proteomics in preclinical drug discovery. Chem. Biol. 2012, 19, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Olsen, R.W.; Huang, J. Identification of direct protein targets of small molecules. ACS Chem. Biol. 2011, 6, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Dejonghe, W.; Russinova, E. Target identification strategies in plant chemical biology. Front. Plant Sci. 2014, 5, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, G.M.; Tucker, C.L.; Xu, T.; Park, S.K.; Han, X.; Yates, J.R., 3rd; Fitzgerald, M.C. Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc. Natl. Acad. Sci. USA 2010, 107, 9078–9082. [Google Scholar] [CrossRef] [PubMed]
- West, G.M.; Tang, L.; Fitzgerald, M.C. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Anal. Chem. 2008, 80, 4175–4185. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wang, D.; Gooden, D.M.; Ball, C.H.; Fitzgerald, M.C. Targeted mass spectrometry-based approach for protein-ligand binding analyses in complex biological mixtures using a phenacyl bromide modification strategy. Anal. Chem. 2016, 88, 10987–10993. [Google Scholar] [CrossRef] [PubMed]
- Saxena, C. Identification of protein binding partners of small molecules using label-free methods. Expert Opin. Drug Discov. 2016, 11, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Strickland, E.C.; Geer, M.A.; Hong, J.; Fitzgerald, M.C. False-positive rate determination of protein target discovery using a covalent modification- and mass spectrometry-based proteomics platform. J. Am. Soc. Mass Spectrom. 2014, 25, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Dearmond, P.D.; Xu, Y.; Strickland, E.C.; Daniels, K.G.; Fitzgerald, M.C. Thermodynamic analysis of protein-ligand interactions in complex biological mixtures using a shotgun proteomics approach. J. Proteome Res. 2011, 10, 4948–4958. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.T.; Adhikari, J.; Fitzgerald, M.C. Stableisotope labeling with amino acids in cell culture (SILAC)-based strategy for proteome-wide thermodynamic analysis of protein-ligand binding interactions. Mol. Cell. Proteom. 2014, 13, 1800–1813. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Wisniewski, J.R.; Cox, J.; Zanivan, S.; Kruger, M.; Ishihama, Y.; Mann, M. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 2011, 6, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Hoedt, E.; Zhang, G.; Neubert, T.A. Stable isotope labeling by amino acids in cell culture (SILAC) for quantitative proteomics. Adv. Exp. Med. Biol. 2014, 806, 93–106. [Google Scholar] [PubMed]
- Larance, M.; Bailly, A.P.; Pourkarimi, E.; Hay, R.T.; Buchanan, G.; Coulthurst, S.; Xirodimas, D.P.; Gartner, A.; Lamond, A.I. Stable-isotope labeling with amino acids in nematodes. Nat. Methods 2011, 8, 849–851. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.E.; Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jin, J.; Zhang, L.; Hu, R.; Gao, L.; Huo, X.; Liu, D.; Ma, X.; Wang, C.; Han, J.; et al. Quantitative analysis of differential protein expression in cervical carcinoma cells after zeylenone treatment by stable isotope labeling with amino acids in cell culture. J. Proteom. 2015, 126, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Martinez Molina, D.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Schirle, M.; Jenkins, J.L. Identifying compound efficacy targets in phenotypic drug discovery. Drug Discov. Today 2016, 21, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Duggan, S.; Richardson, P.L.; Marin, V.; Warder, S.E.; McLoughlin, S.M. Target identification of compounds from a cell viability phenotypic screen using a bead/lysate-based affinity capture platform. J. Biomol. Screen. 2016, 21, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.V.; Olek, K.M.; Muller, A.C.; Tan, C.S.; Bennett, K.L.; Colinge, J.; Superti-Furga, G. Proteome-wide drug and metabolite interaction mapping by thermal-stability profiling. Nat. Methods 2015, 12, 1055–1057. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, F.B.; Eberhard, D.; Werner, T.; Franken, H.; Childs, D.; Doce, C.; Savitski, M.F.; Huber, W.; Bantscheff, M.; Savitski, M.M.; et al. Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nat. Methods 2015, 12, 1129–1131. [Google Scholar] [CrossRef] [PubMed]
- Savitski, M.M.; Reinhard, F.B.; Franken, H.; Werner, T.; Savitski, M.F.; Eberhard, D.; Martinez Molina, D.; Jafari, R.; Dovega, R.B.; Klaeger, S.; et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014, 346, 1255784. [Google Scholar] [CrossRef] [PubMed]
- Becher, I.; Werner, T.; Doce, C.; Zaal, E.A.; Togel, I.; Khan, C.A.; Rueger, A.; Muelbaier, M.; Salzer, E.; Berkers, C.R.; et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 2016, 12, 908–910. [Google Scholar] [CrossRef] [PubMed]
- Mateus, A.; Maatta, T.A.; Savitski, M.M. Thermal proteome profiling: Unbiased assessment of protein state through heat-induced stability changes. Proteome Sci. 2016, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Magtanong, L.; Barker, S.L.; Gresham, D.; Nishimura, S.; Natarajan, P.; Koh, J.L.Y.; Porter, J.; Gray, C.A.; Andersen, R.J.; et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 2009, 27, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Kwon, H.J. Target deconvolution of bioactive small molecules: The heart of chemical biology and drug discovery. Arch. Pharm. Res. 2015, 38, 1627–1641. [Google Scholar] [CrossRef] [PubMed]
- Bassik, M.C.; Kampmann, M.; Lebbink, R.J.; Wang, S.; Hein, M.Y.; Poser, I.; Weibezahn, J.; Horlbeck, M.A.; Chen, S.; Mann, M.; et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 2013, 152, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale crispr-cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Kasap, C.; Elemento, O.; Kapoor, T.M. Drugtargetseqr: A genomics- and CRISPR-Cas9-based method to analyze drug targets. Nat. Chem. Biol. 2014, 10, 626–628. [Google Scholar] [CrossRef] [PubMed]
- Ipsaro, J.J.; Shen, C.; Arai, E.; Xu, Y.; Kinney, J.B.; Joshua-Tor, L.; Vakoc, C.R.; Shi, J. Rapid generation of drug-resistance alleles at endogenous loci using CRISPR-Cas9 indel mutagenesis. PLoS ONE 2017, 12, e0172177. [Google Scholar] [CrossRef] [PubMed]
- Neggers, J.E.; Vercruysse, T.; Jacquemyn, M.; Vanstreels, E.; Baloglu, E.; Shacham, S.; Crochiere, M.; Landesman, Y.; Daelemans, D. Identifying drug-target selectivity of small-molecule CRM1/XPO1 inhibitors by CRISPR/Cas9 genome editing. Chem. Biol. 2015, 22, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Locasale, J.W. Metabolomics: A primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 7216. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Ousterout, D.G.; Gersbach, C.A. Advances in targeted genome editing. Curr. Opin. Chem. Biol. 2012, 16, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Siminovitch, L. Genetic manipulation: Now is the time to consider controls. Sci. Forum 1973, 6, 7–11. [Google Scholar] [PubMed]
- Yarmush, M.L.; Banta, S. Metabolic engineering: Advances in modeling and intervention in health and disease. Ann. Rev. Biomed. Eng. 2003, 5, 349–381. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Fu, Q.; Wang, J.; Ma, S. UPLC-MS/MS determination of ephedrine, methylephedrine, amygdalin and glycyrrhizic acid in beagle plasma and its application to a pharmacokinetic study after oral administration of Ma Huang Tang. Drug. Test. Anal. 2015, 7, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Ekow Thomford, N.; Dzobo, K.; Adu, F.; Chirikure, S.; Wonkam, A.; Dandara, C. Bush mint (Hyptis suaveolens) and spreading hogweed (Boerhavia diffusa) medicinal plant extracts differentially affect activities of CYP1A2, CYP2D6 and CYP3A4 enzymes. J. Ethnopharmacol. 2018, 211, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Plumb, R.; Su, M.; Xu, Z.; Zhao, A.; Qiu, M.; Long, X.; Liu, Z.; Jia, W. Ultra-performance LC/TOF MS analysis of medicinal Panax herbs for metabolomic research. J. Sep. Sci. 2008, 31, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-W.; In, G.; Kim, J.-H.; Cho, B.-G.; Han, G.-H.; Chang, I.-M. Metabolomic approach for discrimination of processed ginseng genus (Panax ginseng and Panax quinquefolius) using UPLC-QTOF MS. J. Ginseng Res. 2014, 38, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Korotcov, A.; Tkachenko, V.; Russo, D.P.; Ekins, S. Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol. Pharm. 2017, 14, 4462–4475. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Matter, H. Integrating virtual screening in lead discovery. Curr. Opin. Chem. Biol. 2004, 8, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Eberherr, C.; Hagemann, M.; Cairo, S.; Haberle, B.; Vokuhl, C.; von Schweinitz, D.; Kappler, R. Connectivity map identifies HDAC inhibition as a treatment option of high-risk hepatoblastoma. Cancer Biol. Ther. 2016, 17, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- Brum, A.M.; van de Peppel, J.; van der Leije, C.S.; Schreuders-Koedam, M.; Eijken, M.; van der Eerden, B.C.; van Leeuwen, J.P. Connectivity map-based discovery of parbendazole reveals targetable human osteogenic pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 12711–12716. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, L.; Kumar, V.; Agarwal, P. Systematic evaluation of connectivity map for disease indications. Genome Med. 2014, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J. The connectivity map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.K.; Berchuck, J.E.; Ross, K.N.; Kakoza, R.M.; Clauser, K.; Schinzel, A.C.; Ross, L.; Galinsky, I.; Davis, T.N.; Silver, S.J.; et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell 2009, 16, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.; Sandhu, S.S.; Sharma, A.K. Prognostic and predictive biomarkers in cancer. Curr. Cancer Drug Targets 2014, 14, 477–504. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, R. Druggable cancer secretome: Neoplasm-associated traits. Cancer Genom. Proteom. 2015, 12, 119–131. [Google Scholar]
- Roti, G.; Stegmaier, K. Genetic and proteomic approaches to identify cancer drug targets. Br. J. Cancer 2012, 106, 254–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, M.; Wright, G.L., Jr.; Hanash, S.M.; Gopal-Srivastava, R.; Srivastava, S. Proteomic approaches within the NCI early detection research network for the discovery and identification of cancer biomarkers. Ann. N. Y. Acad. Sci. 2001, 945, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Awale, M.; Visini, R.; Probst, D.; Arus-Pous, J.; Reymond, J.L. Chemical space: Big data challenge for molecular diversity. Chimia 2017, 71, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Van Driest, S.L.; Wei, W.Q.; Roden, D.M. The influence of big (clinical) data and genomics on precision medicine and drug development. Clin. Pharmacol. Ther. 2018, 103, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Schulthess, D.; Hughes, N.; Vannieuwenhuyse, B.; Kalra, D. Real world big data for clinical research and drug development. Drug Discov. Today 2018, 23, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Kruger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. Chembl: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrian-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Kruger, F.A.; Rostom, R.; Overington, J.P. Mapping small molecule binding data to structural domains. BMC Bioinform. 2012, 13, S11. [Google Scholar]
- Roos, D.S. Computational biology. Bioinformatics—Trying to swim in a sea of data. Science 2001, 291, 1260–1261. [Google Scholar] [CrossRef] [PubMed]
- Jennings, A.; Tennant, M. Discovery strategies in a pharmaceutical setting: The application of computational techniques. Expert Opin. Drug Discov. 2006, 1, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Chen, F. Identifying targets for drug discovery using bioinformatics. Expert Opin. Ther. Targets 2008, 12, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.R.; Xiong, H.; Bengtsson, H.; Bourgon, R.; Gentleman, R. Querying genomic databases: Refining the connectivity map. Stat. Appl. Genet. Mol. Biol. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.S.; Goossens, N.; Hoshida, Y. Use of big data in drug development for precision medicine. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Cappon, G.D. Nonclinical support of pediatric drug development in a global context: An industry perspective. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2011, 92, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Cooper, C.; Mdluli, K. Challenges and opportunities in developing novel drugs for TB. Future Med. Chem. 2011, 3, 1373–1400. [Google Scholar] [CrossRef] [PubMed]
- Morford, L.L.; Bowman, C.J.; Blanset, D.L.; Bogh, I.B.; Chellman, G.J.; Halpern, W.G.; Weinbauer, G.F.; Coogan, T.P. Preclinical safety evaluations supporting pediatric drug development with biopharmaceuticals: Strategy, challenges, current practices. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2011, 92, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Beggs, N.F.; Dobrovolny, H.M. Determining drug efficacy parameters for mathematical models of influenza. J. Biol. Dyn. 2015, 9, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.; Choi, J.; Kwon, M.; Lee, D. Context-specific functional module based drug efficacy prediction. BMC Bioinform. 2016, 17 (Suppl. 6). [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Diaz, M.B.; Viera, S.; Fernandez-Alvaro, E.; Angulo-Barturen, I. Animal models of efficacy to accelerate drug discovery in malaria. Parasitology 2014, 141, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.R.; Johnson, T.; Warren, L.; Hughes, A.R.; Chissoe, S.L.; Xu, C.F.; Waterworth, D.M. The genetics of drug efficacy: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, J.; Onozawa, R.; Fukae, J.; Mishima, T.; Fujioka, S.; Tsuboi, Y. Impact of insufficient drug efficacy of antiparkinson agents on patient’s quality of life: A cross-sectional study. BMC Neurol. 2015, 15, 105. [Google Scholar] [CrossRef] [PubMed]
- Gange, S.J.; Golub, E.T. From smallpox to big data: The next 100 years of epidemiologic methods. Am. J. Epidemiol. 2016, 183, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Docherty, A.B.; Lone, N.I. Exploiting big data for critical care research. Curr. Opin. Crit. Care 2015, 21, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.S.; Tan, J.; Ung, M.; Moore, J.H.; Cheng, C. Big data bioinformatics. J. Cell. Physiol. 2014, 229, 1896–1900. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.S.; Gao, G.; Koch, S. Big data and analytics in healthcare. Methods Inf. Med. 2015, 54, 546–547. [Google Scholar] [CrossRef] [PubMed]
- Wasser, T.; Haynes, K.; Barron, J.; Cziraky, M. Using ‘big data’ to validate claims made in the pharmaceutical approval process. J. Med. Econ. 2015, 18, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Reshef, D.N.; Reshef, Y.A.; Finucane, H.K.; Grossman, S.R.; McVean, G.; Turnbaugh, P.J.; Lander, E.S.; Mitzenmacher, M.; Sabeti, P.C. Detecting novel associations in large data sets. Science 2011, 334, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Omberg, L.; Ellrott, K.; Yuan, Y.; Kandoth, C.; Wong, C.; Kellen, M.R.; Friend, S.H.; Stuart, J.; Liang, H.; Margolin, A.A. Enabling transparent and collaborative computational analysis of 12 tumor types within the cancer genome atlas. Nat. Genet. 2013, 45, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Mias, G.I.; Li-Pook-Than, J.; Jiang, L.; Lam, H.Y.; Chen, R.; Miriami, E.; Karczewski, K.J.; Hariharan, M.; Dewey, F.E.; et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Chapman, T. Lab automation and robotics: Automation on the move. Nature 2003, 421, 661–666. [Google Scholar] [CrossRef] [PubMed]
- King, R.D.; Rowland, J.; Oliver, S.G.; Young, M.; Aubrey, W.; Byrne, E.; Liakata, M.; Markham, M.; Pir, P.; Soldatova, L.N.; et al. The automation of science. Science 2009, 324, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, A.; Aubrey, W.; Byrne, E.; Clare, A.; Khan, M.N.; Liakata, M.; Markham, M.; Rowland, J.; Soldatova, L.N.; Whelan, K.E.; et al. Towards robot scientists for autonomous scientific discovery. Autom. Exp. 2010, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Meanwell, N.A. Improving drug design: An update on recent applications of efficiency metrics, strategies for replacing problematic elements, and compounds in non-traditional drug space. Chem. Res. Toxicol. 2016, 29, 564–616. [Google Scholar] [CrossRef] [PubMed]
- MacConnell, A.B.; Price, A.K.; Paegel, B.M. An integrated microfluidic processor for DNA-encoded combinatorial library functional screening. ACS Comb. Sci. 2017, 19, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Baranczak, A.; Tu, N.P.; Marjanovic, J.; Searle, P.A.; Vasudevan, A.; Djuric, S.W. Integrated platform for expedited synthesis-purification-testing of small molecule libraries. ACS Med. Chem. Lett. 2017, 8, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Muller, A.T.; Huisman, B.J.H.; Fuchs, J.A.; Schneider, P.; Schneider, G. Generative recurrent networks for de novo drug design. Mol. Inform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Merk, D.; Friedrich, L.; Grisoni, F.; Schneider, G. De novo design of bioactive small molecules by artificial intelligence. Mol. Inform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef] [PubMed]
- Duch, W.; Swaminathan, K.; Meller, J. Artificial intelligence approaches for rational drug design and discovery. Curr. Pharm. Des. 2007, 13, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Esch, E.W.; Bahinski, A.; Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015, 14, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Eglen, R.M.; Randle, D.H. Drug discovery goes three-dimensional: Goodbye to flat high-throughput screening? Assay Drug Dev. Technol. 2015, 13, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, V.; Patrinos, G.P. David bowie and the art of slow innovation: A fast-second winner strategy for biotechnology and precision medicine global development. Omics 2017, 21, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.D.; Lalic, G. Teaching target-oriented and diversity-oriented organic synthesis at Harvard University. Chem. Biol. 2002, 9, 535–541. [Google Scholar] [CrossRef]
- Schreiber, S.L. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 2000, 287, 1964–1969. [Google Scholar] [CrossRef] [PubMed]
- Maier, M.E. Design and synthesis of analogues of natural products. Org. Biomol. Chem. 2015, 13, 5302–5343. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Ellinger, B.; Rizzo, S.; Deraeve, C.; Schurmann, M.; Preut, H.; Arndt, H.D.; Waldmann, H. Biology-oriented synthesis of a natural-product inspired oxepane collection yields a small-molecule activator of the Wnt-pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 6805–6810. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, M.; Wetzel, S.; Kumar, K.; Waldmann, H. Biology-inspired synthesis of compound libraries. Cell. Mol. Life Sci. 2008, 65, 1186–1201. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, S.; Bon, R.S.; Kumar, K.; Waldmann, H. Biology-oriented synthesis. Angew. Chem. Int. Ed. Engl. 2011, 50, 10800–10826. [Google Scholar] [CrossRef] [PubMed]
- Wender, P.A.; Quiroz, R.V.; Stevens, M.C. Function through synthesis-informed design. Acc. Chem. Res. 2015, 48, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Wender, P.A.; Verma, V.A.; Paxton, T.J.; Pillow, T.H. Function-oriented synthesis, step economy, and drug design. Acc. Chem. Res. 2008, 41, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Sun, Y.; Challa, S.; Ding, Y.; Lajiness, M.S.; Wild, D.J. Semantic inference using chemogenomics data for drug discovery. BMC Bioinform. 2011, 12, 256. [Google Scholar] [CrossRef] [PubMed]
- White, D.; Wilson, R.C. Generative models for chemical structures. J. Chem. Inf. Model. 2010, 50, 1257–1274. [Google Scholar] [CrossRef] [PubMed]
- Rubio, D.M.; Schoenbaum, E.E.; Lee, L.S.; Schteingart, D.E.; Marantz, P.R.; Anderson, K.E.; Platt, L.D.; Baez, A.; Esposito, K. Defining translational research: Implications for training. Acad. Med. 2010, 85, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Bian, Y.; Hu, Z.; Wang, L.; Xie, X.S. Deep learning for drug design: An artificial intelligence paradigm for drug discovery in the big data era. AAPS J. 2018, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Reutlinger, M.; Rodrigues, T.; Schneider, P.; Schneider, G. Combining on-chip synthesis of a focused combinatorial library with computational target prediction reveals imidazopyridine GPCR ligands. Angew. Chem. Int. Ed. Engl. 2014, 53, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 2018, 17, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Reutlinger, M.; Rodrigues, T.; Schneider, P.; Schneider, G. Multi-objective molecular de novo design by adaptive fragment prioritization. Angew. Chem. Int. Ed. Engl. 2014, 53, 4244–4248. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Rothlisberger, M.; Reker, D.; Schneider, G. Spotting and designing promiscuous ligands for drug discovery. Chem. Commun. 2016, 52, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Y.; Deng, Y.; Kim, B.; Pierce, L.; Krilov, G.; Lupyan, D.; Robinson, S.; Dahlgren, M.K.; Greenwood, J.; et al. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J. Am. Chem. Soc. 2015, 137, 2695–2703. [Google Scholar] [CrossRef] [PubMed]
- Besnard, J.; Ruda, G.F.; Setola, V.; Abecassis, K.; Rodriguiz, R.M.; Huang, X.P.; Norval, S.; Sassano, M.F.; Shin, A.I.; Webster, L.A.; et al. Automated design of ligands to polypharmacological profiles. Nature 2012, 492, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Koppitz, M.; Eis, K. Automated medicinal chemistry. Drug Discov. Today 2006, 11, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D.; Tu, N.P.; Nemcek, T.A.; Searle, P.A.; Hochlowski, J.E.; Djuric, S.W.; Pan, J.Y. An automated synthesis-purification-sample-management platform for the accelerated generation of pharmaceutical candidates. J. Lab. Autom. 2014, 19, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, A.G.; Masquelin, T.; Hemmerle, H. A remote-controlled adaptive medchem lab: An innovative approach to enable drug discovery in the 21st century. Drug Discov. Today 2013, 18, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, C.A.; Watson, I.A.; Hu, H.; Wang, J. The proximal lilly collection: Mapping, exploring and exploiting feasible chemical space. J. Chem. Inf. Model. 2016, 56, 1253–1266. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ballmer, S.G.; Gillis, E.P.; Fujii, S.; Schmidt, M.J.; Palazzolo, A.M.; Lehmann, J.W.; Morehouse, G.F.; Burke, M.D. Synthesis of many different types of organic small molecules using one automated process. Science 2015, 347, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Stalder, R.; Roth, G.P. Preparative microfluidic electrosynthesis of drug metabolites. ACS Med. Chem. Lett. 2013, 4, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Genovino, J.; Sames, D.; Hamann, L.G.; Toure, B.B. Accessing drug metabolites via transition-metal catalyzed c-h oxidation: The liver as synthetic inspiration. Angew. Chem. Int. Ed. Engl. 2016, 55, 14218–14238. [Google Scholar] [CrossRef] [PubMed]
- LaPorte, T.L.; Wang, C. Continuous processes for the production of pharmaceutical intermediates and active pharmaceutical ingredients. Curr. Opin. Drug Discov. Dev. 2007, 10, 738–745. [Google Scholar]
- Chin, P.; Barney, W.S.; Pindzola, B.A. Microstructured reactors as tools for the intensification of pharmaceutical reactions and processes. Curr. Opin. Drug Discov. Dev. 2009, 12, 848–861. [Google Scholar]
- Saaby, S.; Knudsen, K.R.; Ladlow, M.; Ley, S.V. The use of a continuous flow-reactor employing a mixed hydrogen-liquid flow stream for the efficient reduction of imines to amines. Chem. Commun. 2005, 2909–2911. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, M.; O’Brien, M.; Ley, S.V.; Polyzos, A. Flow chemistry: Intelligent processing of gas-liquid transformations using a tube-in-tube reactor. Acc. Chem. Res. 2015, 48, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Loskill, P.; Sezhian, T.; Tharp, K.M.; Lee-Montiel, F.T.; Jeeawoody, S.; Reese, W.M.; Zushin, P.H.; Stahl, A.; Healy, K.E. Wat-on-a-chip: A physiologically relevant microfluidic system incorporating white adipose tissue. Lab Chip 2017, 17, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Eyer, K.; Stratz, S.; Kuhn, P.; Kuster, S.K.; Dittrich, P.S. Implementing enzyme-linked immunosorbent assays on a microfluidic chip to quantify intracellular molecules in single cells. Anal. Chem. 2013, 85, 3280–3287. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M. Frontiers in cancer nanomedicine: Directing mass transport through biological barriers. Trends Biotechnol. 2010, 28, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Zhang, Y.N.; Zhang, W. Cancer-on-a-chip systems at the frontier of nanomedicine. Drug Discov. Today 2017, 22, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Galler, K.; Brautigam, K.; Grosse, C.; Popp, J.; Neugebauer, U. Making a big thing of a small cell–recent advances in single cell analysis. Analyst 2014, 139, 1237–1273. [Google Scholar] [CrossRef] [PubMed]
- Kayala, M.A.; Azencott, C.A.; Chen, J.H.; Baldi, P. Learning to predict chemical reactions. J. Chem. Inf. Model. 2011, 51, 2209–2222. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.R.; Muggleton, S.H.; Sternberg, M.J. Incorporating virtual reactions into a logic-based ligand-based virtual screening method to discover new leads. Mol. Inform. 2015, 34, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Kowalik, M.; Gothard, C.M.; Drews, A.M.; Gothard, N.A.; Weckiewicz, A.; Fuller, P.E.; Grzybowski, B.A.; Bishop, K.J. Parallel optimization of synthetic pathways within the network of organic chemistry. Angew. Chem. Int. Ed. Engl. 2012, 51, 7928–7932. [Google Scholar] [CrossRef] [PubMed]
- Szymkuc, S.; Gajewska, E.P.; Klucznik, T.; Molga, K.; Dittwald, P.; Startek, M.; Bajczyk, M.; Grzybowski, B.A. Computer-assisted synthetic planning: The end of the beginning. Angew. Chem. Int. Ed. Engl. 2016, 55, 5904–5937. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Europe bets on drug discovery. Nature 2013, 494, 20. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rubio, E.; Elizondo, D.A.; Grootveld, M.; Jerez, J.M.; Luque-Baena, R.M. Computational intelligence techniques in medicine. Comput. Math. Methods Med. 2015, 2015, 196976. [Google Scholar] [CrossRef] [PubMed]
- Montanez-Godinez, N.; Martinez-Olguin, A.C.; Deeb, O.; Garduno-Juarez, R.; Ramirez-Galicia, G. QSAR/QSPR as an application of artificial neural networks. Methods Mol. Biol. 2015, 1260, 319–333. [Google Scholar] [PubMed]
- Wesolowski, M.; Suchacz, B. Artificial neural networks: Theoretical background and pharmaceutical applications: A review. J. AOAC Int. 2012, 95, 652–668. [Google Scholar] [CrossRef] [PubMed]
- Baskin, I.I.; Winkler, D.; Tetko, I.V. A renaissance of neural networks in drug discovery. Expert Opin. Drug Discov. 2016, 11, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Nikolsky, Y.; Nikolskaya, T.; Bugrim, A. Biological networks and analysis of experimental data in drug discovery. Drug Discov. Today 2005, 10, 653–662. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Congreve, M.; Carr, R.; Murray, C.; Jhoti, H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov. Today 2003, 8, 876–877. [Google Scholar] [CrossRef]
- Van Molle, I.; Thomann, A.; Buckley, D.L.; So, E.C.; Lang, S.; Crews, C.M.; Ciulli, A. Dissecting fragment-based lead discovery at the von Hippel-Lindau protein: Hypoxia inducible factor 1alpha protein-protein interface. Chem. Biol. 2012, 19, 1300–1312. [Google Scholar] [CrossRef] [PubMed]
- Zuegg, J.; Cooper, M.A. Drug-likeness and increased hydrophobicity of commercially available compound libraries for drug screening. Curr. Top. Med. Chem. 2012, 12, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Lifongo, L.L.; Judson, P.N.; Sippl, W.; Efange, S.M. How “drug-like” are naturally occurring anti-cancer compounds? J. Mol. Model. 2014, 20, 2069. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, K.; Baringhaus, K.H.; Schneider, G. Scaffold diversity of natural products: Inspiration for combinatorial library design. Nat. Prod. Rep. 2008, 25, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Elumalai, N.; Berg, A.; Natarajan, K.; Scharow, A.; Berg, T. Nanomolar inhibitors of the transcription factor STAT5b with high selectivity over STAT5a. Angew. Chem. Int. Ed. Engl. 2015, 54, 4758–4763. [Google Scholar] [CrossRef] [PubMed]
- Bon, R.S.; Waldmann, H. Bioactivity-guided navigation of chemical space. Acc. Chem. Res. 2010, 43, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.; van Otterlo, W.A.; Dominguez Seoane, M.; Mocklinghoff, S.; Hofmann, B.; Wetzel, S.; Schuffenhauer, A.; Ertl, P.; Oprea, T.I.; Steinhilber, D.; et al. Bioactivity-guided mapping and navigation of chemical space. Nat. Chem. Biol. 2009, 5, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, S.; Klein, K.; Renner, S.; Rauh, D.; Oprea, T.I.; Mutzel, P.; Waldmann, H. Interactive exploration of chemical space with scaffold hunter. Nat. Chem. Biol. 2009, 5, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Reker, D.; Schneider, P.; Schneider, G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. Pass: Prediction of activity spectra for biologically active substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Stepanchikova, A.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. Prediction of biological activity spectra for substances: Evaluation on the diverse sets of drug-like structures. Curr. Med. Chem. 2003, 10, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Reker, D.; Rodrigues, T.; Schneider, P.; Schneider, G. Identifying the macromolecular targets of de novo-designed chemical entities through self-organizing map consensus. Proc. Natl. Acad. Sci. USA 2014, 111, 4067–4072. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Reker, D.; Rodrigues, T.; Schneider, P. Coping with polypharmacology by computational medicinal chemistry. Chimia 2014, 68, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed]
- DiMasi, J.A.; Feldman, L.; Seckler, A.; Wilson, A. Trends in risks associated with new drug development: Success rates for investigational drugs. Clin. Pharmacol. Ther. 2010, 87, 272–277. [Google Scholar] [CrossRef] [PubMed]
- DiMasi, J.A.; Reichert, J.M.; Feldman, L.; Malins, A. Clinical approval success rates for investigational cancer drugs. Clin. Pharmacol. Ther. 2013, 94, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Loong, H.H.; Siu, L.L. Selecting the best drugs for phase I clinical development and beyond. In American Society of Clinical Oncology Educational Book. American Society of Clinical Oncology. Meeting; American Society of Clinical Oncology: Alexandria, VA, USA, 2013; pp. 469–473. [Google Scholar]
- Chavan, S.; Nicholls, I.A.; Karlsson, B.C.; Rosengren, A.M.; Ballabio, D.; Consonni, V.; Todeschini, R. Towards global qsar model building for acute toxicity: Munro database case study. Int. J. Mol. Sci. 2014, 15, 18162–18174. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. Qsar modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed]
- Devillers, J. Methods for building QSARs. Methods Mol. Biol. 2013, 930, 3–27. [Google Scholar] [PubMed]
- Sullivan, K.M.; Manuppello, J.R.; Willett, C.E. Building on a solid foundation: SAR and QSAR as a fundamental strategy to reduce animal testing. SAR QSAR Environ. Res. 2014, 25, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Kirchmair, J.; Goller, A.H.; Lang, D.; Kunze, J.; Testa, B.; Wilson, I.D.; Glen, R.C.; Schneider, G. Predicting drug metabolism: Experiment and/or computation? Nat. Rev. Drug Discov. 2015, 14, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Lal Gupta, P.; Jayaram, B. Predicting the binding modes and sites of metabolism of xenobiotics. Mol. BioSyst. 2015, 11, 1914–1924. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Min, J.L.; Lin, W.Z.; Liu, Z.; Cheng, X.; Chou, K.C. Idrug-target: Predicting the interactions between drug compounds and target proteins in cellular networking via benchmark dataset optimization approach. J. Biomol. Struct. Dyn. 2015, 33, 2221–2233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, F.; Luo, L.; Zhang, J. Predicting drug side effects by multi-label learning and ensemble learning. BMC Bioinform. 2015, 16, 365. [Google Scholar] [CrossRef] [PubMed]
- Bastian, L.; Hof, J.; Pfau, M.; Fichtner, I.; Eckert, C.; Henze, G.; Prada, J.; von Stackelberg, A.; Seeger, K.; Shalapour, S. Synergistic activity of bortezomib and hdaci in preclinical models of b-cell precursor acute lymphoblastic leukemia via modulation of p53, pi3k/akt, and nf-kappab. Clin. Cancer Res. 2013, 19, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
- Stanciu-Herrera, C.; Morgan, C.; Herrera, L. Anti-cd19 and anti-cd22 monoclonal antibodies increase the effectiveness of chemotherapy in pre-b acute lymphoblastic leukemia cell lines. Leukemia Res. 2008, 32, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Senthebane, D.A.; Rowe, A.; Thomford, N.E.; Mwapagha, L.M.; Al-Awwad, N.; Dandara, C.; Parker, M.I. Cancer stem cell hypothesis for therapeutic innovation in clinical oncology? Taking the root out, not chopping the leaf. Omics 2016, 20, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Senthebane, D.A.; Thomford, N.E.; Rowe, A.; Dandara, C.; Parker, M.I. Not everyone fits the mold: Intratumor and intertumor heterogeneity and innovative cancer drug design and development. Omics 2018, 22, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Cortes, J.; Kim, S.B.; Im, S.A.; Hegg, R.; Im, Y.H.; Roman, L.; Pedrini, J.L.; Pienkowski, T.; Knott, A.; et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 2012, 366, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Kawajiri, H.; Takashima, T.; Kashiwagi, S.; Noda, S.; Onoda, N.; Hirakawa, K. Pertuzumab in combination with trastuzumab and docetaxel for HER2-positive metastatic breast cancer. Expert Rev. Anticancer Ther. 2015, 15, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Baselga, J.; Kim, S.B.; Ro, J.; Semiglazov, V.; Campone, M.; Ciruelos, E.; Ferrero, J.M.; Schneeweiss, A.; Heeson, S.; et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N. Engl. J. Med. 2015, 372, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Baselga, J.; Miles, D.; Im, Y.H.; Quah, C.; Lee, L.F.; Cortes, J. Incidence of central nervous system metastases in patients with HER2-positive metastatic breast cancer treated with pertuzumab, trastuzumab, and docetaxel: Results from the randomized phase iii study cleopatra. Ann. Oncol. 2014, 25, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Kim, S.B.; Cortes, J.; Ro, J.; Semiglazov, V.; Campone, M.; Ciruelos, E.; Ferrero, J.M.; Schneeweiss, A.; Knott, A.; et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (cleopatra study): Overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2013, 14, 461–471. [Google Scholar] [CrossRef]
- National Academy of Sciences (US). The national academies collection: Reports funded by national institutes of health. In Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease; National Academies Press (US), National Academy of Sciences: Washington, DC, USA, 2011. [Google Scholar]
- Debouck, C. Integrating genomics across drug discovery and development. Toxicol. Lett. 2009, 186, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Debouck, C.; Metcalf, B. The impact of genomics on drug discovery. Ann. Rev. Pharmacol. Toxicol. 2000, 40, 193–207. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. Int. J. Mol. Sci. 2018, 19, 1578. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19061578
Thomford NE, Senthebane DA, Rowe A, Munro D, Seele P, Maroyi A, Dzobo K. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. International Journal of Molecular Sciences. 2018; 19(6):1578. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19061578
Chicago/Turabian StyleThomford, Nicholas Ekow, Dimakatso Alice Senthebane, Arielle Rowe, Daniella Munro, Palesa Seele, Alfred Maroyi, and Kevin Dzobo. 2018. "Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery" International Journal of Molecular Sciences 19, no. 6: 1578. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19061578