The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients
Abstract
:1. Introduction
2. Results
2.1. Frequency of CMV DNA Detection in Saliva and Plasma
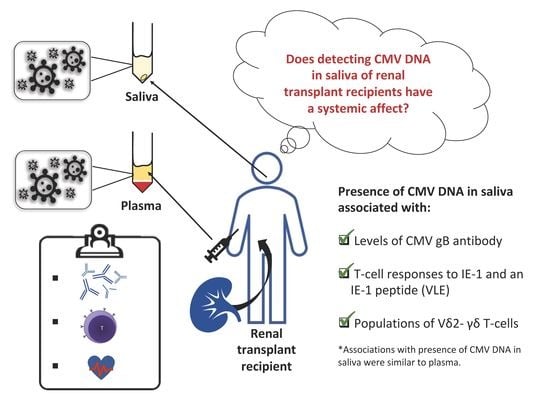
2.2. CMV DNA Detected in Saliva is Associated with Immunological Responses to CMV
2.3. CMV DNA Displayed Weak Positive Associations with Cardiovascular Risk
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Detection of CMV DNA in Plasma and Saliva
4.3. Immunological Assessments of CMV Burden
4.4. Assessment of Vascular Pathology
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AU | Arbitrary units |
| cIMT | Carotid intimal media thickness |
| CMV | Human cytomegalovirus |
| CVD | Cardiovascular disease |
| FMD | Flow mediated dilatation |
| PBMC | Peripheral blood mononuclear cells |
| qPCR | Quantitative PCR |
| RTR | Renal transplant recipients |
References
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Peng, G.; Bai, J.; He, B.; Huang, K.; Hu, X.; Liu, D. Cytomegalovirus Infection and Relative Risk of Cardiovascular Disease (Ischemic Heart Disease, Stroke, and Cardiovascular Death): A Meta-Analysis of Prospective Studies Up to 2016. J. Am. Heart Assoc. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jovana, C.; Sladjana, U.; Ivan, J.; Milan, V.; Vladisav, S. Viral infection in renal transplant recipients. Sci. World J. 2012, 2012, 820621. [Google Scholar]
- Fischer, S.A.; Avery, R.K.; AST Infectious Disease Community of Practice. Screening of donor and recipient prior to solid organ transplantation. Am. J. Transplant. 2009, 9, S7–S18. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.T.; Haan, M.N.; Dowd, J.B.; Aiello, A.E. Cytomegalovirus antibody levels, inflammation, and mortality among elderly Latinos over 9 years of follow-up. Am. J. Epidemiol. 2010, 172, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Lichtner, M.; Cicconi, P.; Vita, S.; Cozzi-Lepri, A.; Galli, M.; Lo Caputo, S.; Saracino, A.; De Luca, A.; Moioli, M.; Maggiolo, F.; et al. Cytomegalovirus coinfection is associated with an increased risk of severe non-AIDS-defining events in a large cohort of HIV-infected patients. J. Infect. Dis. 2015, 211, 178–186. [Google Scholar] [CrossRef]
- Karim, B.; Wijaya, I.P.; Rahmaniy, R.; Ariyanto, I.; Waters, S.; Estiasari, R.; Price, P. Factors affecting affect cardiovascular health in Indonesian HIV patients beginning ART. AIDS Res. Ther. 2017, 14, 52. [Google Scholar] [CrossRef]
- Affandi, J.S.; Montgomery, J.; Brunt, S.J.; Nolan, D.; Price, P. The immunological footprint of CMV in HIV-1 patients stable on long-term ART. Immun. Ageing 2015, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Shariff, N.; Cobbold, M.; Bruton, R.; Ainsworth, J.A.; Sinclair, A.J.; Nayak, L.; Moss, P.A. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol. 2002, 169, 1984–1992. [Google Scholar] [CrossRef]
- Waters, S.; Brook, E.; Lee, S.; Estiasari, R.; Ariyanto, I.; Price, P. HIV patients, healthy aging and transplant recipients can reveal the hidden footprints of CMV. Clin. Immunol. 2018, 187, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Groh, V.; Porcelli, S.; Fabbi, M.; Lanier, L.L.; Picker, L.J.; Anderson, T.; Warnke, R.A.; Bhan, A.K.; Strominger, J.L.; Brenner, M.B. Human lymphocytes bearing T cell receptor gamma/delta are phenotypically diverse and evenly distributed throughout the lymphoid system. J. Exp. Med. 1989, 169, 1277–1294. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.R.; Elliott, L.; Hussey, S.; Mahmud, N.; Kelly, J.; Doherty, D.G.; Feighery, C.F. Persistent changes in circulating and intestinal gammadelta T cell subsets, invariant natural killer T cells and mucosal-associated invariant T cells in children and adults with coeliac disease. PLoS ONE 2013, 8, e76008. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Affandi, J.S.; Irish, A.B.; Price, P. Cytomegalovirus infection alters phenotypes of different gammadelta T-cell subsets in renal transplant recipients with long-term stable graft function. J. Med. Virol. 2017, 89, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Razonable, R.R.; Humar, A. Cytomegalovirus in solid organ transplant recipients-Guidelines of the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant. 2019, 33, e13512. [Google Scholar] [CrossRef] [PubMed]
- Koidl, C.; Bozic, M.; Marth, E.; Kessler, H.H. Detection of CMV DNA: Is EDTA whole blood superior to EDTA plasma? J. Virol. Methods 2008, 154, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Von Muller, L.; Hinz, J.; Bommer, M.; Hampl, W.; Kluwick, S.; Wiedmann, M.; Bunjes, D.; Mertens, T. CMV monitoring using blood cells and plasma: A comparison of apples with oranges? Bone Marrow Transplant. 2007, 39, 353–357. [Google Scholar] [CrossRef]
- Von Muller, L.; Hampl, W.; Hinz, J.; Meisel, H.; Reip, A.; Engelmann, E.; Heilbronn, R.; Gartner, B.; Kramer, O.; Einsele, H.; et al. High variability between results of different in-house tests for cytomegalovirus (CMV) monitoring and a standardized quantitative plasma CMV PCR assay. J. Clin. Microbiol. 2002, 40, 2285–2287. [Google Scholar] [CrossRef]
- Neonatal Directorate Management Committee. Cytomegalovirus (CMV) Neonatal Pathway. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=2&ved=2ahUKEwiH8aGjkaXlAhVtHKYKHTL8DuMQFjABegQIARAC&url=https%3A%2F%2Fwww.kemh.health.wa.gov.au%2F~%2Fmedia%2FFiles%2FHospitals%2FWNHS%2FFor%2520health%2520professionals%2FClinical%2520guidelines%2FNEO%2FWNHS.NEO.CytomegalovirusCMVNeonatalPathway.pdf&usg=AOvVaw2J7Zoi-qFtQUq9CmNiHCkw (accessed on 22 November 2016).
- Amin, M.M.; Bialek, S.R.; Dollard, S.C.; Wang, C. Urinary Cytomegalovirus Shedding in the United States: The National Health and Nutrition Examination Surveys, 1999–2004. Clin. Infect. Dis. 2018, 67, 587–592. [Google Scholar] [CrossRef]
- Morrison, K.M.; Beucler, M.J.; Campbell, E.O.; White, M.A.; Boody, R.E.; Wilson, K.C.; Miller, W.E. Development of a Primary Human Cell Model for the Study of Human Cytomegalovirus Replication and Spread within Salivary Epithelium. J. Virol. 2019. [Google Scholar] [CrossRef]
- Brantsaeter, A.B.; Holberg-Petersen, M.; Jeansson, S.; Goplen, A.K.; Bruun, J.N. CMV quantitative PCR in the diagnosis of CMV disease in patients with HIV-infection-a retrospective autopsy based study. BMC Infect. Dis. 2007, 7, 127. [Google Scholar] [CrossRef]
- Britt, W.J.; Vugler, L.; Butfiloski, E.J.; Stephens, E.B. Cell surface expression of human cytomegalovirus (HCMV) gp55–116 (gB): Use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J. Virol. 1990, 64, 1079–1085. [Google Scholar] [PubMed]
- Gonczol, E.; deTaisne, C.; Hirka, G.; Berencsi, K.; Lin, W.C.; Paoletti, E.; Plotkin, S. High expression of human cytomegalovirus (HCMV)-gB protein in cells infected with a vaccinia-gB recombinant: The importance of the gB protein in HCMV immunity. Vaccine 1991, 9, 631–637. [Google Scholar] [CrossRef]
- Pitard, V.; Roumanes, D.; Lafarge, X.; Couzi, L.; Garrigue, I.; Lafon, M.E.; Merville, P.; Moreau, J.F.; Dechanet-Merville, J. Long-term expansion of effector/memory Vdelta2-gammadelta T cells is a specific blood signature of CMV infection. Blood 2008, 112, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Nickel, P.; Bold, G.; Presber, F.; Biti, D.; Babel, N.; Kreutzer, S.; Pratschke, J.; Schonemann, C.; Kern, F.; Volk, H.D.; et al. High levels of CMV-IE-1-specific memory T cells are associated with less alloimmunity and improved renal allograft function. Transpl. Immunol. 2009, 20, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Leng, S.X.; Qu, T.; Semba, R.D.; Li, H.; Yao, X.; Nilles, T.; Yang, X.; Manwani, B.; Walston, J.D.; Ferrucci, L.; et al. Relationship between cytomegalovirus (CMV) IgG serology, detectable CMV DNA in peripheral monocytes, and CMV pp65(495–503)-specific CD8+ T cells in older adults. Age 2011, 33, 607–614. [Google Scholar] [CrossRef]
- Lee, S.; Brook, E.; Affandi, J.; Howson, P.; Tanudjaja, S.A.; Dhaliwal, S.; Irish, A.; Price, P. A high burden of cytomegalovirus marks poor vascular health in transplant recipients more clearly than in the general population. Clin. Transl. Immunol. 2019, 8, e1043. [Google Scholar] [CrossRef] [Green Version]
- Raitakari, O.T.; Celermajer, D.S. Flow-mediated dilatation. Br. J. Clin. Pharmacol. 2000, 50, 397–404. [Google Scholar] [CrossRef]
- O’Brien, K.D.; Allen, M.D.; McDonald, T.O.; Chait, A.; Harlan, J.M.; Fishbein, D.; McCarty, J.; Ferguson, M.; Hudkins, K.; Benjamin, C.D.; et al. Vascular cell adhesion molecule-1 is expressed in human coronary atherosclerotic plaques. Implications for the mode of progression of advanced coronary atherosclerosis. J. Clin. Investig. 1993, 92, 945–951. [Google Scholar] [CrossRef]
- O’Brien, K.D.; McDonald, T.O.; Chait, A.; Allen, M.D.; Alpers, C.E. Neovascular expression of E-selectin, intercellular adhesion molecule-1, and vascular cell adhesion molecule-1 in human atherosclerosis and their relation to intimal leukocyte content. Circulation 1996, 93, 672–682. [Google Scholar] [CrossRef]
- Molenaar, T.J.; Twisk, J.; de Haas, S.A.; Peterse, N.; Vogelaar, B.J.; van Leeuwen, S.H.; Michon, I.N.; van Berkel, T.J.; Kuiper, J.; Biessen, E.A. P-selectin as a candidate target in atherosclerosis. Biochem. Pharmacol. 2003, 66, 859–866. [Google Scholar] [CrossRef]
- Bath, P.M.; May, J.; Heptinstall, S. Clinical utility of remote platelet function measurement using P-selectin: Assessment of aspirin, clopidogrel, and prasugrel and bleeding disorders. Platelets 2018, 29, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Price, P.; Lee, S.; Affandi, J.; Parsons, R.; Naylor, L.H.; Watts, G.F.; Irish, A. Cytomegalovirus antibody and vascular pathology in renal transplant recipients. J. Med. Virol. 2017, 89, 177–181. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waters, S.; Lee, S.; Lloyd, M.; Irish, A.; Price, P. The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients. Int. J. Mol. Sci. 2019, 20, 5230. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205230
Waters S, Lee S, Lloyd M, Irish A, Price P. The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients. International Journal of Molecular Sciences. 2019; 20(20):5230. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205230
Chicago/Turabian StyleWaters, Shelley, Silvia Lee, Megan Lloyd, Ashley Irish, and Patricia Price. 2019. "The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients" International Journal of Molecular Sciences 20, no. 20: 5230. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205230