Published online Oct 10, 2015. doi: 10.5306/wjco.v6.i5.92
Peer-review started: May 29, 2015
First decision: June 18, 2015
Revised: July 8, 2015
Accepted: July 29, 2015
Article in press: August 3, 2015
Published online: October 10, 2015
Alternative splicing (AS), the process of removing introns from pre-mRNA and re-arrangement of exons to give several types of mature transcripts, has been described more than 40 years ago. However, until recently, it has not been clear how extensive it is. Genome-wide studies have now conclusively shown that more than 90% of genes are alternatively spliced in humans. This makes AS one of the main drivers of proteomic diversity and, consequently, determinant of cellular function repertoire. Unsurprisingly, given its extent, numerous splice isoforms have been described to be associated with several diseases including cancer. Many of them have antagonistic functions, e.g., pro- and anti-angiogenic or pro- and anti-apoptotic. Additionally several splice factors have been recently described to have oncogene or tumour suppressors activities, like SF3B1 which is frequently mutated in myelodysplastic syndromes. Beside the implications for cancer pathogenesis, de-regulated AS is recognized as one of the novel areas of cell biology where therapeutic manipulations may be designed. This editorial discusses the possibilities of manipulation of AS for therapeutic benefit in cancer. Approaches involving the use of oligonucleotides as well as small molecule splicing modulators are presented as well as thoughts on how specificity might be accomplished in splicing therapeutics.
Core tip: Genome-wide studies have recently shown that more than 90% of genes are alternatively spliced in humans. This makes alternative splicing (AS) one of the main drivers of proteomic diversity. Numerous splice isoforms have been described to be associated with cancer. Additionally several splice factors have been shown to have oncogene or tumour suppressors activities. Beside the implications for cancer pathogenesis, de-regulated AS is recognized as one of the novel areas of cell biology where therapeutic manipulations may be designed. This editorial discusses the possibilities of manipulation of AS for therapeutic benefit in cancer.
- Citation: Oltean S. Modulators of alternative splicing as novel therapeutics in cancer. World J Clin Oncol 2015; 6(5): 92-95
- URL: https://www.wjgnet.com/2218-4333/full/v6/i5/92.htm
- DOI: https://dx.doi.org/10.5306/wjco.v6.i5.92
In the last years we have seen a plethora of anticancer agents that try to acquire more specific and targeted treatment in comparison with the conventional chemo- and radiotherapies used in the clinic. While it is highly unlikely they will be able to be used as mono-therapies on a large scale in oncology - due to the inherent problem of developing resistant clones as exemplified by the B-Raf inhibitor vemurafenib in melanoma[1], they have certainly proved very useful in combination therapies or as adjuvants that can improve overall survival in association with conventional therapies or reduce the doses used in chemo- and radiotherapies and therefore decrease side-effects.
Most of targeted anti-cancer drugs approved in clinical practice today are targeting receptor tyrosine kinases or cytoplasmic signalling molecules. However, since cancer cells are different from normal cells in virtually any property and function from DNA repair to regulating apoptosis or metabolism, theoretically drugs that hamper tumour growth may be designed at any level of gene regulation – transcriptional, post-transcriptional or post-translational. Indeed, recent years have produced intense research on potential new drugs (some already in trials or in the clinic) that are based on epigenetic modulation[2], DNA repair[3] or microRNAs[4] to name a few.
One level that has not been explored so far is represented by modulation of alternative splicing (AS).
Splicing is the removal of introns during processing of pre-mRNA. Through AS the composition of the mature RNA may be changed through exon skipping, mutually exclusive exons, intron retention or 3’ and 5’ alternative splice sites[5]. AS has emerged in the post-genomic era as the main driver of proteome diversity with at least 94% of multi-exon genes being alternatively spliced in humans[6,7]. AS is one of the main control mechanisms for cell phenotype, and a process deregulated in disease. There are over 2000 splicing mutations known, involving 303 genes and implicated in 370 diseases[8]. Therefore it has become essential to study how this process is regulated, and how it can become deregulated in disease.
While the disease most commonly linked to deregulation of AS in several genes is cancer[9], there are many in-depth reports of pathogenic splice variants in diseases ranging from neuromuscular disorders[10] to diabetes[11] or cardiomyopathies[12].
An increasing amount of literature in the last years shows involvement of splicing in cancer and an incredible number of splice variants have been described to be associated with tumour progression - for recent reviews see[9,13,14]. For example, epidermal growth factor receptor, which is mutated in several cancers, has a splice variant that is missing exon 4 and is highly expressed in several cancers; this exon deletion makes the protein constitutively active[15]. K-Ras has two alternate exons - 4A and 4B - and depending on their inclusion/exclusion there is a strong differential association with various forms or localization of colon cancer[16]. The tumour suppressor p53 has two splice isoforms p53beta and p53gamma that result from two alternate exons; these isoforms modulate the activity of the main isoform and the way it regulates apoptosis in various contexts[17]. Finally, another notable example is the well-studied tumour suppressor retinoblastoma protein for which more than 15% of the mutations described in various cancers are related to splicing[18,19].
The main question that arises - especially having a therapeutic purpose in mind - are these modifications simply by-products of the oncogenic process or do they drive pathogenesis of cancer? While inevitably some splice variants are “associated noise” similar to physiology, there is compelling evidence for “pathogenic” AS in cancer.
Firstly, similar with mutations in transcription factors that denote many of them as oncogenes, there are mutations of spliceosome components or splice factors - e.g., SF3B1 in myelodysplastic syndromes[20].
Secondly, there is clear evidence of splicing-specific variants that may be induced by signalling in the cancer cell environment and result in acquired functions for the cancer cells that helps their pathogenic evolution. For example, while normal cells/tissues generally have a high level of the anti-angiogenic vascular endothelial growth factor A (VEGF-A) isoforms VEGF165b, this is lost in cancers, with expression of predominantly pro-angiogenic VEGF165a, which maintains a state of high and chaotic neovascularization in tumours[21]. However, no mutation has been identified so far that could account for this shift in the ratio of the two splice isoforms which is highly likely due to changes in the microenvironment during step-wise progression of the oncogenic process.
Finally, recent years have clearly shown that defective splicing contributes to one of the most challenging problems in oncology - acquired resistance to treatments. While there are numerous examples[22] we want to point-out the well-known case of Vemurafenib. Patients treated with this drug invariably develop resistance. While several mechanisms have been described, in about a third of cases this occurs through faulty AS that results in truncated B-Raf which do not have the Ras-binding domain[23].
Can we modify splicing and use it as a new level where therapeutic interventions may be designed? While there is no drug in the clinic that modifies splicing yet, there are certainly extremely exciting developments in the past few years. The general idea is to try and switch the splicing of a certain isoform that has been identified as deleterious and promoting the oncogenic process in functional studies towards a beneficial isoform.
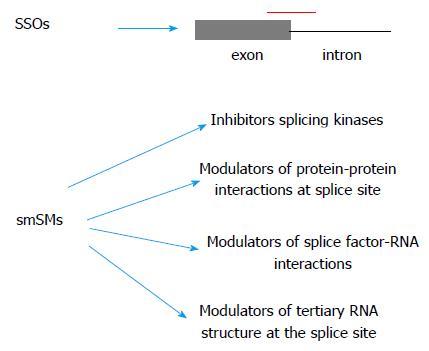
The strategy most used so far involves anti-sense oligos (ASO) or splicing-switching oligos (SSOs). The general principle is to design ASOs that bind either exon-intron junctions or regulatory sequences like enhancers or silencers in introns or exons, therefore affecting the splice outcome of the targeted event. So far SSOs have been proved very promising, with several of them in clinical trials, e.g., for Duchenne muscular dystrophy or spinal muscular atrophy[24].
There is a growing number of small-molecule splicing modulators (smSM) that have been shown to affect splicing. An interesting example is amiloride. This is a long-time used diuretic with the main mechanism of action through effects on the ion pumps in the renal tubules. However, it has been found in a screen to potently affect splicing of several genes involved in apoptosis and further-on to be able to decrease tumour growth in animal models[25]. Recently a class of small molecule compounds that inhibit SRPK1, a major regulator of AS through SR-protein phosphorylation, has been shown to inhibit VEGF splicing and angiogenesis in a model of ocular neovascularization[26] as well as melanoma xenografts growth[27] and orthotopic prostate cancer mouse models[28].
Potentially, other types of molecules could be involved in splicing modulation, like chemicals that affect splice factor/RNA interactions or molecules that affect directly the tertiary structure of a particular splice junction (Figure 1).
Specificity is highly unlikely to be an important problem for SSOs, which are designed to bind on defined RNA sequences, though potential problems with delivery and toxicity might still be challenging.
SmSMs could potentially affect several other splice events regulated by the same splicing kinase or splicing factor intended to be modulating - however, the key issue is whether the manipulation of the intended targeted splice event is dominant functionally in the system/cell line of interest (i.e., the other splice events affected do not result in major unintended modifications in cell properties).
It is interesting to point-out a recent paper reporting the development of smSMs of the SMN splicing and attenuation of spinal muscular atrophy[10]. The compounds were found in a screen using a splicing reporter that mimicked the endogenous splicing event. When an RNA-seq analysis was performed to assess specificity it was found that very few splice junctions are affected, therefore proving that specificity in splicing therapeutics using small molecules may be accomplished.
P- Reviewer: Arcaini L, Cheung L, Su CC S- Editor: Tian YL L- Editor: A E- Editor: Jiao XK
| 1. | Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085-3096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 785] [Cited by in F6Publishing: 758] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 2. | West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 943] [Cited by in F6Publishing: 1021] [Article Influence: 102.1] [Reference Citation Analysis (0)] |
| 3. | Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30:326-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 188] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 4. | Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121-1126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 241] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 5. | Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 909] [Cited by in F6Publishing: 930] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 6. | Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-1415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2521] [Cited by in F6Publishing: 2631] [Article Influence: 164.4] [Reference Citation Analysis (0)] |
| 7. | Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3539] [Cited by in F6Publishing: 3662] [Article Influence: 228.9] [Reference Citation Analysis (0)] |
| 8. | Wang J, Zhang J, Li K, Zhao W, Cui Q. SpliceDisease database: linking RNA splicing and disease. Nucleic Acids Res. 2012;40:D1055-D1059. [PubMed] [Cited in This Article: ] |
| 9. | Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311-5318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 414] [Cited by in F6Publishing: 448] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 10. | Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KK, Karp GM, Qi H, Woll MG. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688-693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 346] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 11. | Le Bacquer O, Shu L, Marchand M, Neve B, Paroni F, Kerr Conte J, Pattou F, Froguel P, Maedler K. TCF7L2 splice variants have distinct effects on beta-cell turnover and function. Hum Mol Genet. 2011;20:1906-1915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 12. | Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766-773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 369] [Cited by in F6Publishing: 390] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 13. | Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol. 2014;32:30-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 14. | Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 15. | Wang H, Zhou M, Shi B, Zhang Q, Jiang H, Sun Y, Liu J, Zhou K, Yao M, Gu J. Identification of an exon 4-deletion variant of epidermal growth factor receptor with increased metastasis-promoting capacity. Neoplasia. 2011;13:461-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 16. | Abubaker J, Bavi P, Al-Haqawi W, Sultana M, Al-Harbi S, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435-445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Marcel V, Fernandes K, Terrier O, Lane DP, Bourdon JC. Modulation of p53β and p53γ expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ. 2014;21:1377-1387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 18. | Zhang K, Nowak I, Rushlow D, Gallie BL, Lohmann DR. Patterns of missplicing caused by RB1 gene mutations in patients with retinoblastoma and association with phenotypic expression. Hum Mutat. 2008;29:475-484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Lohmann DR. RB1 gene mutations in retinoblastoma. Hum Mutat. 1999;14:283-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 20. | Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384-1395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 917] [Cited by in F6Publishing: 928] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 21. | Harper SJ, Bates DO. VEGF-A splicing: the key to anti-angiogenic therapeutics? Nat Rev Cancer. 2008;8:880-887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 22. | Eblen ST. Regulation of chemoresistance via alternative messenger RNA splicing. Biochem Pharmacol. 2012;83:1063-1072. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Salton M, Kasprzak WK, Voss T, Shapiro BA, Poulikakos PI, Misteli T. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat Commun. 2015;6:7103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 24. | Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472-482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 325] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 25. | Ding Y, Zhang H, Zhou Z, Zhong M, Chen Q, Wang X, Zhu Z. u-PA inhibitor amiloride suppresses peritoneal metastasis in gastric cancer. World J Surg Oncol. 2012;10:270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Gammons MV, Fedorov O, Ivison D, Du C, Clark T, Hopkins C, Hagiwara M, Dick AD, Cox R, Harper SJ. Topical antiangiogenic SRPK1 inhibitors reduce choroidal neovascularization in rodent models of exudative AMD. Invest Ophthalmol Vis Sci. 2013;54:6052-6062. [PubMed] [Cited in This Article: ] |
| 27. | Gammons MV, Lucas R, Dean R, Coupland SE, Oltean S, Bates DO. Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma. Br J Cancer. 2014;111:477-485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 28. | Mavrou A, Brakspear K, Hamdollah-Zadeh M, Damodaran G, Babaei-Jadidi R, Oxley J, Gillatt DA, Ladomery MR, Harper SJ, Bates DO. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene. 2015;34:4311-4319. [PubMed] [Cited in This Article: ] |